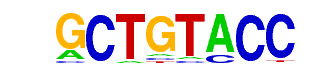
| p-value: | 1e-14 |
| log p-value: | -3.261e+01 |
| Information Content per bp: | 1.810 |
| Number of Target Sequences with motif | 20.0 |
| Percentage of Target Sequences with motif | 3.32% |
| Number of Background Sequences with motif | 2.8 |
| Percentage of Background Sequences with motif | 0.42% |
| Average Position of motif in Targets | 21.3 +/- 11.4bp |
| Average Position of motif in Background | 8.5 +/- 1.9bp |
| Strand Bias (log2 ratio + to - strand density) | 1.0 |
| Multiplicity (# of sites on avg that occur together) | 1.06 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer
| Match Rank: | 1 |
| Score: | 0.75
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GCTGTACC---
-CTGTTCCTGG |
|


|
|
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 2 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCTGTACC
GCTGTG-- |
|

|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GCTGTACC
NGCTN---- |
|


|
|
PH0158.1_Rhox11_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GCTGTACC----
AGGACGCTGTAAAGGGA |
|

|
|
MA0499.1_Myod1/Jaspar
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GCTGTACC-
TGCAGCTGTCCCT |
|


|
|
PH0157.1_Rhox11_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GCTGTACC----
AAGACGCTGTAAAGCGA |
|

|
|
PB0156.1_Plagl1_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GCTGTACC-------
NNNNGGTACCCCCCANN |
|


|
|
MA0007.2_AR/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------GCTGTACC-
GNACANNNTGTTCTT |
|


|
|
MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GCTGTACC
CAGCTGTT-- |
|


|
|
Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GCTGTACC
BAACAGCTGT--- |
|


|
|