







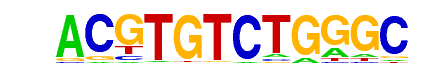











| p-value: | 1e-1436 |
| log p-value: | -3.307e+03 |
| Information Content per bp: | 1.587 |
| Number of Target Sequences with motif | 1402.0 |
| Percentage of Target Sequences with motif | 36.08% |
| Number of Background Sequences with motif | 46.6 |
| Percentage of Background Sequences with motif | 1.61% |
| Average Position of motif in Targets | 24.8 +/- 10.7bp |
| Average Position of motif in Background | 29.1 +/- 20.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.16 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.902 |  | 1e-1011 | -2330.079078 | 42.82% | 5.43% | motif file (matrix) |
| 2 | 0.886 |  | 1e-914 | -2105.781829 | 43.03% | 6.36% | motif file (matrix) |
| 3 | 0.859 |  | 1e-867 | -1998.181500 | 50.33% | 10.15% | motif file (matrix) |
| 4 | 0.854 |  | 1e-783 | -1803.959901 | 59.93% | 17.03% | motif file (matrix) |
| 5 | 0.840 |  | 1e-541 | -1245.809791 | 10.63% | 0.23% | motif file (matrix) |
| 6 | 0.756 |  | 1e-450 | -1037.563583 | 11.43% | 0.48% | motif file (matrix) |
| 7 | 0.642 |  | 1e-283 | -652.970000 | 7.18% | 0.29% | motif file (matrix) |
| 8 | 0.668 |  | 1e-272 | -626.348046 | 5.76% | 0.15% | motif file (matrix) |
| 9 | 0.672 |  | 1e-196 | -452.607778 | 3.24% | 0.05% | motif file (matrix) |
| 10 | 0.643 |  | 1e-125 | -289.222947 | 3.04% | 0.13% | motif file (matrix) |
| 11 | 0.668 |  | 1e-121 | -279.493827 | 5.07% | 0.55% | motif file (matrix) |
| 12 | 0.705 |  | 1e-119 | -275.176373 | 5.28% | 0.59% | motif file (matrix) |
| 13 | 0.604 |  | 1e-110 | -254.091879 | 2.47% | 0.08% | motif file (matrix) |
| 14 | 0.692 |  | 1e-101 | -233.508535 | 4.30% | 0.47% | motif file (matrix) |
| 15 | 0.662 |  | 1e-93 | -215.183963 | 3.24% | 0.26% | motif file (matrix) |
| 16 | 0.617 |  | 1e-81 | -188.212984 | 4.12% | 0.57% | motif file (matrix) |
| 17 | 0.646 |  | 1e-37 | -86.762174 | 1.96% | 0.28% | motif file (matrix) |
| 18 | 0.615 |  | 1e-34 | -79.001253 | 1.08% | 0.10% | motif file (matrix) |