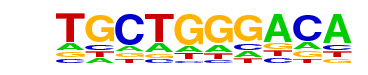
| p-value: | 1e-56 |
| log p-value: | -1.310e+02 |
| Information Content per bp: | 1.448 |
| Number of Target Sequences with motif | 81.0 |
| Percentage of Target Sequences with motif | 2.08% |
| Number of Background Sequences with motif | 5.9 |
| Percentage of Background Sequences with motif | 0.20% |
| Average Position of motif in Targets | 25.7 +/- 12.7bp |
| Average Position of motif in Background | 29.6 +/- 14.2bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGCTGGGACA-
HTGCTGAGTCAT |
|


|
|
AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TGCTGGGACA-
-CCAGGAACAG |
|


|
|
NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCTGGGACA--
TGCTGAGTCATC |
|


|
|
MA0591.1_Bach1::Mafk/Jaspar
| Match Rank: | 4 |
| Score: | 0.70
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGCTGGGACA----
NTGCTGAGTCATCCN |
|


|
|
Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 5 |
| Score: | 0.70
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----TGCTGGGACA-
AWWNTGCTGAGTCAT |
|

|
|
ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer
| Match Rank: | 6 |
| Score: | 0.69
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TGCTGGGACA-
RGSMTBCTGGGAAAT |
|


|
|
MA0501.1_NFE2::MAF/Jaspar
| Match Rank: | 7 |
| Score: | 0.69
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TGCTGGGACA-
AAANTGCTGAGTCAT |
|

|
|
MA0150.2_Nfe2l2/Jaspar
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCTGGGACA-----
TGCTGAGTCATNNTG |
|


|
|
MA0144.2_STAT3/Jaspar
| Match Rank: | 9 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGCTGGGACA
CTTCTGGGAAA |
|


|
|
PB0206.1_Zic2_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.66
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TGCTGGGACA
TCNCCTGCTGNGNNN |
|


|
|