





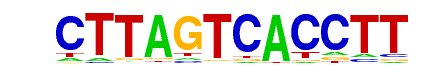



| p-value: | 1e-1761 |
| log p-value: | -4.055e+03 |
| Information Content per bp: | 1.623 |
| Number of Target Sequences with motif | 3178.0 |
| Percentage of Target Sequences with motif | 10.27% |
| Number of Background Sequences with motif | 316.6 |
| Percentage of Background Sequences with motif | 1.24% |
| Average Position of motif in Targets | 25.3 +/- 12.2bp |
| Average Position of motif in Background | 25.6 +/- 14.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.996 |  | 1e-1622 | -3736.518293 | 10.32% | 1.41% | motif file (matrix) |
| 2 | 0.974 |  | 1e-1580 | -3639.656637 | 13.04% | 2.47% | motif file (matrix) |
| 3 | 0.912 |  | 1e-1222 | -2814.582396 | 12.13% | 2.76% | motif file (matrix) |
| 4 | 0.765 |  | 1e-154 | -355.292613 | 0.90% | 0.11% | motif file (matrix) |
| 5 | 0.630 |  | 1e-118 | -273.136291 | 0.62% | 0.07% | motif file (matrix) |
| 6 | 0.761 |  | 1e-69 | -160.479124 | 1.10% | 0.35% | motif file (matrix) |
| 7 | 0.635 |  | 1e-66 | -152.429914 | 0.24% | 0.01% | motif file (matrix) |
| 8 | 0.768 |  | 1e-46 | -106.201997 | 0.34% | 0.06% | motif file (matrix) |