





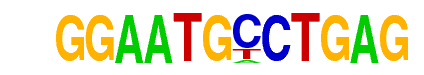










| p-value: | 1e-190 |
| log p-value: | -4.391e+02 |
| Information Content per bp: | 1.639 |
| Number of Target Sequences with motif | 1818.0 |
| Percentage of Target Sequences with motif | 5.87% |
| Number of Background Sequences with motif | 695.5 |
| Percentage of Background Sequences with motif | 2.73% |
| Average Position of motif in Targets | 25.1 +/- 12.1bp |
| Average Position of motif in Background | 24.5 +/- 12.6bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
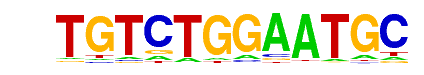
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.901 |  | 1e-175 | -403.554051 | 3.52% | 1.31% | motif file (matrix) |
| 2 | 0.648 |  | 1e-156 | -359.893714 | 6.03% | 3.08% | motif file (matrix) |
| 3 | 0.859 |  | 1e-148 | -341.076324 | 0.32% | 0.01% | motif file (matrix) |
| 4 | 0.889 |  | 1e-145 | -334.793669 | 8.14% | 4.73% | motif file (matrix) |
| 5 | 0.613 |  | 1e-141 | -325.001736 | 5.23% | 2.63% | motif file (matrix) |
| 6 | 0.637 |  | 1e-73 | -168.161230 | 8.24% | 5.70% | motif file (matrix) |
| 7 | 0.686 |  | 1e-59 | -137.576653 | 0.22% | 0.01% | motif file (matrix) |
| 8 | 0.708 |  | 1e-54 | -125.078257 | 0.25% | 0.02% | motif file (matrix) |
| 9 | 0.735 |  | 1e-47 | -109.480059 | 0.16% | 0.01% | motif file (matrix) |
| 10 | 0.620 |  | 1e-44 | -103.234453 | 0.27% | 0.04% | motif file (matrix) |
| 11 | 0.644 |  | 1e-43 | -100.921713 | 0.60% | 0.18% | motif file (matrix) |
| 12 | 0.650 |  | 1e-43 | -99.321937 | 1.05% | 0.44% | motif file (matrix) |
| 13 | 0.621 |  | 1e-39 | -91.514826 | 0.15% | 0.01% | motif file (matrix) |
| 14 | 0.786 |  | 1e-31 | -71.940573 | 0.14% | 0.01% | motif file (matrix) |
| 15 | 0.677 |  | 1e-26 | -60.310342 | 0.34% | 0.10% | motif file (matrix) |
| 16 | 0.700 |  | 1e-22 | -52.672414 | 0.12% | 0.01% | motif file (matrix) |