





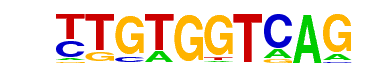





| p-value: | 1e-112 |
| log p-value: | -2.585e+02 |
| Information Content per bp: | 1.569 |
| Number of Target Sequences with motif | 108.0 |
| Percentage of Target Sequences with motif | 9.87% |
| Number of Background Sequences with motif | 2.9 |
| Percentage of Background Sequences with motif | 0.53% |
| Average Position of motif in Targets | 48.0 +/- 26.8bp |
| Average Position of motif in Background | 55.8 +/- 24.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.06 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.954 |  | 1e-97 | -223.642282 | 16.64% | 2.40% | motif file (matrix) |
| 2 | 0.801 |  | 1e-87 | -201.799836 | 9.41% | 0.72% | motif file (matrix) |
| 3 | 0.601 |  | 1e-69 | -160.555517 | 12.80% | 1.97% | motif file (matrix) |
| 4 | 0.638 |  | 1e-63 | -145.575505 | 7.59% | 0.73% | motif file (matrix) |
| 5 | 0.657 |  | 1e-56 | -129.301492 | 8.50% | 0.95% | motif file (matrix) |
| 6 | 0.697 |  | 1e-46 | -106.913710 | 7.59% | 0.95% | motif file (matrix) |
| 7 | 0.721 |  | 1e-40 | -93.694561 | 4.94% | 0.47% | motif file (matrix) |
| 8 | 0.675 |  | 1e-30 | -70.729743 | 4.11% | 0.40% | motif file (matrix) |
| 9 | 0.613 |  | 1e-30 | -69.759040 | 3.20% | 0.33% | motif file (matrix) |
| 10 | 0.705 |  | 1e-24 | -56.455041 | 4.20% | 0.58% | motif file (matrix) |