


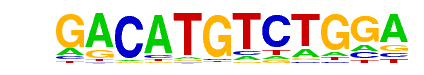


















| p-value: | 1e-2900 |
| log p-value: | -6.679e+03 |
| Information Content per bp: | 1.520 |
| Number of Target Sequences with motif | 1150.0 |
| Percentage of Target Sequences with motif | 24.41% |
| Number of Background Sequences with motif | 1.0 |
| Percentage of Background Sequences with motif | 0.03% |
| Average Position of motif in Targets | 24.7 +/- 8.7bp |
| Average Position of motif in Background | 29.2 +/- 2.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
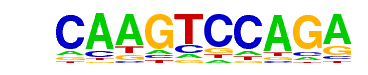
| 1 | 0.914 |  | 1e-2054 | -4730.057849 | 30.81% | 0.54% | motif file (matrix) |
| 2 | 0.855 |  | 1e-1318 | -3036.263630 | 37.95% | 3.39% | motif file (matrix) |
| 3 | 0.823 |  | 1e-1123 | -2587.247238 | 28.82% | 1.95% | motif file (matrix) |
| 4 | 0.789 |  | 1e-1087 | -2503.708903 | 13.94% | 0.14% | motif file (matrix) |
| 5 | 0.787 |  | 1e-1045 | -2408.244034 | 40.81% | 5.99% | motif file (matrix) |
| 6 | 0.742 |  | 1e-958 | -2206.277148 | 11.01% | 0.09% | motif file (matrix) |
| 7 | 0.740 |  | 1e-726 | -1672.559300 | 30.33% | 4.59% | motif file (matrix) |
| 8 | 0.771 |  | 1e-511 | -1178.076003 | 7.30% | 0.10% | motif file (matrix) |
| 9 | 0.696 |  | 1e-478 | -1102.217930 | 11.12% | 0.57% | motif file (matrix) |
| 10 | 0.710 |  | 1e-362 | -833.656089 | 5.60% | 0.11% | motif file (matrix) |
| 11 | 0.704 |  | 1e-336 | -775.265263 | 6.03% | 0.16% | motif file (matrix) |
| 12 | 0.765 |  | 1e-316 | -729.269516 | 3.97% | 0.02% | motif file (matrix) |
| 13 | 0.630 |  | 1e-305 | -703.672103 | 5.90% | 0.21% | motif file (matrix) |
| 14 | 0.613 |  | 1e-256 | -589.738449 | 3.35% | 0.06% | motif file (matrix) |
| 15 | 0.682 |  | 1e-240 | -553.317871 | 5.20% | 0.22% | motif file (matrix) |
| 16 | 0.619 |  | 1e-214 | -493.403797 | 4.58% | 0.20% | motif file (matrix) |
| 17 | 0.648 |  | 1e-186 | -429.819330 | 3.08% | 0.09% | motif file (matrix) |
| 18 | 0.616 |  | 1e-167 | -385.738522 | 3.86% | 0.21% | motif file (matrix) |
| 19 | 0.648 |  | 1e-128 | -296.852275 | 2.84% | 0.13% | motif file (matrix) |
| 20 | 0.681 |  | 1e-108 | -250.208259 | 2.31% | 0.11% | motif file (matrix) |
| 21 | 0.620 |  | 1e-75 | -173.157966 | 2.40% | 0.24% | motif file (matrix) |