| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-100 | -2.316e+02 | 0.0000 | 53.0 | 25.48% | 1.5 | 0.22% | motif file (matrix) |
pdf |
| 2 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-100 | -2.316e+02 | 0.0000 | 53.0 | 25.48% | 1.5 | 0.22% | motif file (matrix) |
pdf |
| 3 |  | p63(p53)/Keratinocyte-p63-ChIP-Seq(GSE17611)/Homer | 1e-63 | -1.472e+02 | 0.0000 | 53.0 | 25.48% | 5.4 | 0.77% | motif file (matrix) |
pdf |
| 4 |  | p53(p53)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-27 | -6.240e+01 | 0.0000 | 27.0 | 12.98% | 4.7 | 0.68% | motif file (matrix) |
pdf |
| 5 |  | Tcfcp2l1(CP2)/mES-Tcfcp2l1-ChIP-Seq(GSE11431)/Homer | 1e-6 | -1.412e+01 | 0.0000 | 6.0 | 2.88% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 6 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.255e+00 | 0.0107 | 4.0 | 1.92% | 1.5 | 0.21% | motif file (matrix) |
pdf |
| 7 | 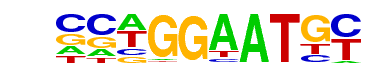 | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-3 | -7.924e+00 | 0.0127 | 13.0 | 6.25% | 14.5 | 2.10% | motif file (matrix) |
pdf |
| 8 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.873e+00 | 0.0127 | 5.0 | 2.40% | 2.4 | 0.35% | motif file (matrix) |
pdf |
| 9 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-2 | -5.715e+00 | 0.0901 | 4.0 | 1.92% | 2.8 | 0.41% | motif file (matrix) |
pdf |
| 10 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.647e+00 | 0.0901 | 9.0 | 4.33% | 10.2 | 1.46% | motif file (matrix) |
pdf |
| 11 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.639e+00 | 0.0901 | 12.0 | 5.77% | 16.6 | 2.39% | motif file (matrix) |
pdf |





















