
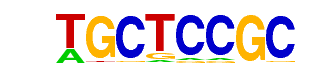



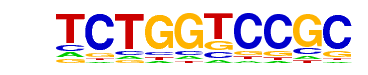




| p-value: | 1e-94 |
| log p-value: | -2.173e+02 |
| Information Content per bp: | 1.627 |
| Number of Target Sequences with motif | 70.0 |
| Percentage of Target Sequences with motif | 24.65% |
| Number of Background Sequences with motif | 2.0 |
| Percentage of Background Sequences with motif | 0.50% |
| Average Position of motif in Targets | 27.5 +/- 12.1bp |
| Average Position of motif in Background | 6.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.9 |
| Multiplicity (# of sites on avg that occur together) | 1.19 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.867 |  | 1e-80 | -185.013494 | 18.66% | 0.19% | motif file (matrix) |
| 2 | 0.607 |  | 1e-64 | -149.466220 | 15.85% | 0.21% | motif file (matrix) |
| 3 | 0.676 |  | 1e-48 | -111.497235 | 12.68% | 0.27% | motif file (matrix) |
| 4 | 0.635 |  | 1e-41 | -95.411404 | 11.27% | 0.40% | motif file (matrix) |
| 5 | 0.687 |  | 1e-41 | -95.411404 | 11.27% | 0.00% | motif file (matrix) |
| 6 | 0.644 |  | 1e-36 | -83.708638 | 10.21% | 0.40% | motif file (matrix) |
| 7 | 0.627 |  | 1e-25 | -57.990773 | 9.51% | 0.72% | motif file (matrix) |
| 8 | 0.604 |  | 1e-23 | -54.261659 | 7.39% | 0.00% | motif file (matrix) |