



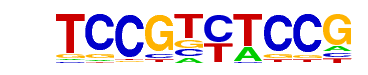




| p-value: | 1e-86 |
| log p-value: | -1.987e+02 |
| Information Content per bp: | 1.623 |
| Number of Target Sequences with motif | 56.0 |
| Percentage of Target Sequences with motif | 19.72% |
| Number of Background Sequences with motif | 1.7 |
| Percentage of Background Sequences with motif | 0.42% |
| Average Position of motif in Targets | 23.2 +/- 13.0bp |
| Average Position of motif in Background | 25.3 +/- 7.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.17 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.812 |  | 1e-82 | -189.564186 | 19.01% | 0.39% | motif file (matrix) |
| 2 | 0.809 |  | 1e-76 | -175.981453 | 17.96% | 0.36% | motif file (matrix) |
| 3 | 0.727 |  | 1e-41 | -95.411404 | 11.27% | 0.33% | motif file (matrix) |
| 4 | 0.603 |  | 1e-39 | -91.474492 | 10.92% | 0.08% | motif file (matrix) |
| 5 | 0.616 |  | 1e-37 | -85.319179 | 14.08% | 0.74% | motif file (matrix) |
| 6 | 0.687 |  | 1e-29 | -68.640014 | 8.80% | 0.15% | motif file (matrix) |
| 7 | 0.707 |  | 1e-21 | -48.957313 | 8.45% | 0.54% | motif file (matrix) |