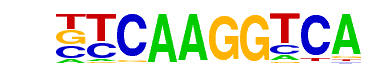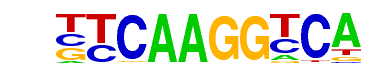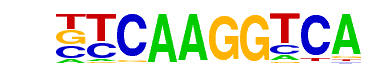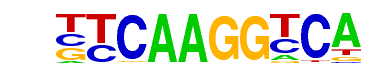| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-51 | -1.197e+02 | 0.0000 | 38.0 | 13.38% | 0.6 | 0.15% | motif file (matrix) |
pdf |
| 2 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-39 | -9.147e+01 | 0.0000 | 31.0 | 10.92% | 0.7 | 0.17% | motif file (matrix) |
pdf |
| 3 | 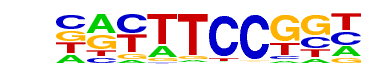 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-28 | -6.609e+01 | 0.0000 | 37.0 | 13.03% | 4.3 | 1.05% | motif file (matrix) |
pdf |
| 4 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-19 | -4.400e+01 | 0.0000 | 18.0 | 6.34% | 0.8 | 0.20% | motif file (matrix) |
pdf |
| 5 |  | ETS(ETS)/Promoter/Homer | 1e-10 | -2.513e+01 | 0.0000 | 12.0 | 4.23% | 0.6 | 0.15% | motif file (matrix) |
pdf |
| 6 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-10 | -2.448e+01 | 0.0000 | 15.0 | 5.28% | 2.1 | 0.52% | motif file (matrix) |
pdf |
| 7 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-9 | -2.289e+01 | 0.0000 | 19.0 | 6.69% | 4.4 | 1.08% | motif file (matrix) |
pdf |
| 8 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-8 | -1.899e+01 | 0.0000 | 17.0 | 5.99% | 4.4 | 1.09% | motif file (matrix) |
pdf |
| 9 | 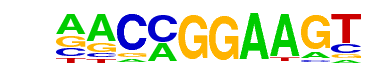 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-5 | -1.176e+01 | 0.0002 | 26.0 | 9.15% | 14.0 | 3.45% | motif file (matrix) |
pdf |
| 10 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-4 | -1.034e+01 | 0.0008 | 12.0 | 4.23% | 4.4 | 1.07% | motif file (matrix) |
pdf |
| 11 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-4 | -9.687e+00 | 0.0014 | 17.0 | 5.99% | 8.4 | 2.08% | motif file (matrix) |
pdf |
| 12 |  | ZNF711(Zf)/SH-SY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-4 | -9.410e+00 | 0.0017 | 29.0 | 10.21% | 19.3 | 4.75% | motif file (matrix) |
pdf |
| 13 |  | E2F(E2F)/Cell-Cycle-Exp/Homer | 1e-4 | -9.360e+00 | 0.0017 | 6.0 | 2.11% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 14 | 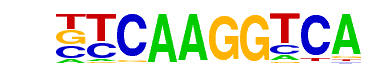 | Nr5a2(NR)/Pancreas-LRH1-ChIP-Seq(GSE34295)/Homer | 1e-3 | -7.178e+00 | 0.0104 | 5.0 | 1.76% | 0.9 | 0.23% | motif file (matrix) |
pdf |
| 15 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-3 | -7.178e+00 | 0.0104 | 5.0 | 1.76% | 2.0 | 0.49% | motif file (matrix) |
pdf |
| 16 | 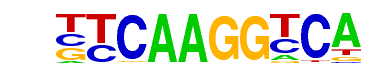 | Nr5a2(NR)/mES-Nr5a2-ChIP-Seq(GSE19019)/Homer | 1e-3 | -7.178e+00 | 0.0104 | 5.0 | 1.76% | 0.8 | 0.20% | motif file (matrix) |
pdf |
| 17 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-3 | -7.178e+00 | 0.0104 | 5.0 | 1.76% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 18 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-3 | -7.178e+00 | 0.0104 | 5.0 | 1.76% | 0.7 | 0.18% | motif file (matrix) |
pdf |
| 19 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-2 | -6.899e+00 | 0.0131 | 23.0 | 8.10% | 16.7 | 4.11% | motif file (matrix) |
pdf |
| 20 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-2 | -5.779e+00 | 0.0362 | 6.0 | 2.11% | 2.4 | 0.58% | motif file (matrix) |
pdf |
| 21 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.779e+00 | 0.0362 | 6.0 | 2.11% | 2.2 | 0.55% | motif file (matrix) |
pdf |
| 22 |  | Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer | 1e-2 | -5.354e+00 | 0.0529 | 21.0 | 7.39% | 16.2 | 3.97% | motif file (matrix) |
pdf |
| 23 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-2 | -5.179e+00 | 0.0577 | 7.0 | 2.46% | 3.2 | 0.78% | motif file (matrix) |
pdf |
| 24 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-2 | -5.179e+00 | 0.0577 | 7.0 | 2.46% | 3.5 | 0.85% | motif file (matrix) |
pdf |
| 25 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -5.174e+00 | 0.0577 | 4.0 | 1.41% | 0.1 | 0.03% | motif file (matrix) |
pdf |
| 26 |  | NRF1/Promoter/Homer | 1e-2 | -5.174e+00 | 0.0577 | 4.0 | 1.41% | 0.4 | 0.10% | motif file (matrix) |
pdf |