| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-981 | -2.259e+03 | 0.0000 | 1860.0 | 42.58% | 231.4 | 6.58% | motif file (matrix) |
pdf |
| 2 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-934 | -2.152e+03 | 0.0000 | 1909.0 | 43.70% | 263.6 | 7.50% | motif file (matrix) |
pdf |
| 3 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-848 | -1.954e+03 | 0.0000 | 1926.0 | 44.09% | 302.8 | 8.61% | motif file (matrix) |
pdf |
| 4 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-719 | -1.658e+03 | 0.0000 | 1017.0 | 23.28% | 72.5 | 2.06% | motif file (matrix) |
pdf |
| 5 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-267 | -6.171e+02 | 0.0000 | 582.0 | 13.32% | 74.5 | 2.12% | motif file (matrix) |
pdf |
| 6 | 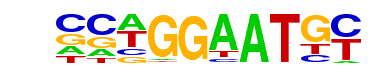 | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-154 | -3.568e+02 | 0.0000 | 1086.0 | 24.86% | 374.9 | 10.66% | motif file (matrix) |
pdf |
| 7 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-149 | -3.435e+02 | 0.0000 | 1007.0 | 23.05% | 338.1 | 9.61% | motif file (matrix) |
pdf |
| 8 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-124 | -2.862e+02 | 0.0000 | 194.0 | 4.44% | 15.6 | 0.44% | motif file (matrix) |
pdf |
| 9 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-99 | -2.280e+02 | 0.0000 | 194.0 | 4.44% | 22.0 | 0.62% | motif file (matrix) |
pdf |
| 10 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-94 | -2.181e+02 | 0.0000 | 375.0 | 8.59% | 85.8 | 2.44% | motif file (matrix) |
pdf |
| 11 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-88 | -2.036e+02 | 0.0000 | 149.0 | 3.41% | 13.8 | 0.39% | motif file (matrix) |
pdf |
| 12 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-68 | -1.584e+02 | 0.0000 | 222.0 | 5.08% | 42.8 | 1.22% | motif file (matrix) |
pdf |
| 13 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-68 | -1.575e+02 | 0.0000 | 248.0 | 5.68% | 52.1 | 1.48% | motif file (matrix) |
pdf |
| 14 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-62 | -1.445e+02 | 0.0000 | 662.0 | 15.16% | 267.9 | 7.62% | motif file (matrix) |
pdf |
| 15 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-58 | -1.348e+02 | 0.0000 | 898.0 | 20.56% | 419.5 | 11.93% | motif file (matrix) |
pdf |
| 16 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-56 | -1.308e+02 | 0.0000 | 1194.0 | 27.34% | 618.4 | 17.59% | motif file (matrix) |
pdf |
| 17 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-56 | -1.290e+02 | 0.0000 | 766.0 | 17.54% | 342.7 | 9.75% | motif file (matrix) |
pdf |
| 18 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-48 | -1.128e+02 | 0.0000 | 997.0 | 22.83% | 507.8 | 14.44% | motif file (matrix) |
pdf |
| 19 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-48 | -1.126e+02 | 0.0000 | 688.0 | 15.75% | 309.7 | 8.81% | motif file (matrix) |
pdf |
| 20 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-45 | -1.036e+02 | 0.0000 | 940.0 | 21.52% | 480.7 | 13.67% | motif file (matrix) |
pdf |
| 21 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-42 | -9.788e+01 | 0.0000 | 705.0 | 16.14% | 334.7 | 9.52% | motif file (matrix) |
pdf |
| 22 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-41 | -9.576e+01 | 0.0000 | 531.0 | 12.16% | 229.6 | 6.53% | motif file (matrix) |
pdf |
| 23 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-37 | -8.633e+01 | 0.0000 | 305.0 | 6.98% | 108.5 | 3.08% | motif file (matrix) |
pdf |
| 24 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-29 | -6.678e+01 | 0.0000 | 1520.0 | 34.80% | 951.0 | 27.04% | motif file (matrix) |
pdf |
| 25 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-28 | -6.632e+01 | 0.0000 | 748.0 | 17.12% | 400.6 | 11.39% | motif file (matrix) |
pdf |
| 26 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-27 | -6.249e+01 | 0.0000 | 870.0 | 19.92% | 489.7 | 13.93% | motif file (matrix) |
pdf |
| 27 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-26 | -6.071e+01 | 0.0000 | 1108.0 | 25.37% | 660.1 | 18.77% | motif file (matrix) |
pdf |
| 28 | 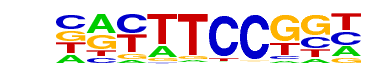 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-25 | -5.915e+01 | 0.0000 | 740.0 | 16.94% | 405.5 | 11.53% | motif file (matrix) |
pdf |
| 29 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-25 | -5.766e+01 | 0.0000 | 526.0 | 12.04% | 265.7 | 7.56% | motif file (matrix) |
pdf |
| 30 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-22 | -5.185e+01 | 0.0000 | 678.0 | 15.52% | 374.9 | 10.66% | motif file (matrix) |
pdf |
| 31 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-22 | -5.122e+01 | 0.0000 | 498.0 | 11.40% | 255.4 | 7.26% | motif file (matrix) |
pdf |
| 32 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-21 | -4.893e+01 | 0.0000 | 536.0 | 12.27% | 283.8 | 8.07% | motif file (matrix) |
pdf |
| 33 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-20 | -4.614e+01 | 0.0000 | 551.0 | 12.61% | 298.0 | 8.47% | motif file (matrix) |
pdf |
| 34 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-19 | -4.553e+01 | 0.0000 | 794.0 | 18.18% | 465.4 | 13.23% | motif file (matrix) |
pdf |
| 35 | 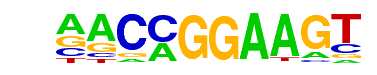 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-19 | -4.444e+01 | 0.0000 | 873.0 | 19.99% | 523.8 | 14.90% | motif file (matrix) |
pdf |
| 36 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-17 | -4.043e+01 | 0.0000 | 604.0 | 13.83% | 342.7 | 9.75% | motif file (matrix) |
pdf |
| 37 |  | Tlx?(NR)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-17 | -3.949e+01 | 0.0000 | 287.0 | 6.57% | 135.4 | 3.85% | motif file (matrix) |
pdf |
| 38 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-16 | -3.912e+01 | 0.0000 | 710.0 | 16.25% | 418.9 | 11.91% | motif file (matrix) |
pdf |
| 39 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-16 | -3.861e+01 | 0.0000 | 737.0 | 16.87% | 438.3 | 12.47% | motif file (matrix) |
pdf |
| 40 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-16 | -3.811e+01 | 0.0000 | 572.0 | 13.10% | 324.4 | 9.22% | motif file (matrix) |
pdf |
| 41 |  | NF1(CTF)/LNCAP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-15 | -3.661e+01 | 0.0000 | 309.0 | 7.07% | 152.8 | 4.35% | motif file (matrix) |
pdf |
| 42 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-15 | -3.636e+01 | 0.0000 | 299.0 | 6.85% | 146.5 | 4.17% | motif file (matrix) |
pdf |
| 43 |  | Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer | 1e-15 | -3.591e+01 | 0.0000 | 279.0 | 6.39% | 134.9 | 3.84% | motif file (matrix) |
pdf |
| 44 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-15 | -3.583e+01 | 0.0000 | 435.0 | 9.96% | 236.0 | 6.71% | motif file (matrix) |
pdf |
| 45 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-15 | -3.556e+01 | 0.0000 | 2645.0 | 60.55% | 1916.6 | 54.51% | motif file (matrix) |
pdf |
| 46 |  | CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-15 | -3.455e+01 | 0.0000 | 573.0 | 13.12% | 331.2 | 9.42% | motif file (matrix) |
pdf |
| 47 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-14 | -3.408e+01 | 0.0000 | 227.0 | 5.20% | 104.5 | 2.97% | motif file (matrix) |
pdf |
| 48 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-14 | -3.346e+01 | 0.0000 | 389.0 | 8.91% | 208.4 | 5.93% | motif file (matrix) |
pdf |
| 49 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-14 | -3.335e+01 | 0.0000 | 577.0 | 13.21% | 336.4 | 9.57% | motif file (matrix) |
pdf |
| 50 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-14 | -3.327e+01 | 0.0000 | 884.0 | 20.24% | 555.5 | 15.80% | motif file (matrix) |
pdf |
| 51 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-14 | -3.314e+01 | 0.0000 | 245.0 | 5.61% | 116.8 | 3.32% | motif file (matrix) |
pdf |
| 52 |  | Mef2a(MADS)/HL1-Mef2a.biotin-ChIP-Seq(GSE21529/Homer | 1e-14 | -3.280e+01 | 0.0000 | 386.0 | 8.84% | 207.3 | 5.90% | motif file (matrix) |
pdf |
| 53 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-14 | -3.225e+01 | 0.0000 | 682.0 | 15.61% | 412.6 | 11.73% | motif file (matrix) |
pdf |
| 54 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-13 | -3.141e+01 | 0.0000 | 206.0 | 4.72% | 94.6 | 2.69% | motif file (matrix) |
pdf |
| 55 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-13 | -3.067e+01 | 0.0000 | 354.0 | 8.10% | 189.7 | 5.39% | motif file (matrix) |
pdf |
| 56 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-12 | -2.892e+01 | 0.0000 | 344.0 | 7.88% | 185.0 | 5.26% | motif file (matrix) |
pdf |
| 57 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-12 | -2.874e+01 | 0.0000 | 1289.0 | 29.51% | 869.7 | 24.73% | motif file (matrix) |
pdf |
| 58 |  | NFkB-p65-Rel(RHD)/LPS-exp(GSE23622)/Homer | 1e-11 | -2.685e+01 | 0.0000 | 73.0 | 1.67% | 23.3 | 0.66% | motif file (matrix) |
pdf |
| 59 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-11 | -2.638e+01 | 0.0000 | 279.0 | 6.39% | 146.2 | 4.16% | motif file (matrix) |
pdf |
| 60 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-11 | -2.622e+01 | 0.0000 | 494.0 | 11.31% | 292.9 | 8.33% | motif file (matrix) |
pdf |
| 61 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-11 | -2.538e+01 | 0.0000 | 634.0 | 14.51% | 393.9 | 11.20% | motif file (matrix) |
pdf |
| 62 |  | Sp1(Zf)/Promoter/Homer | 1e-10 | -2.403e+01 | 0.0000 | 68.0 | 1.56% | 22.2 | 0.63% | motif file (matrix) |
pdf |
| 63 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-9 | -2.235e+01 | 0.0000 | 947.0 | 21.68% | 631.6 | 17.96% | motif file (matrix) |
pdf |
| 64 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-9 | -2.172e+01 | 0.0000 | 931.0 | 21.31% | 621.2 | 17.67% | motif file (matrix) |
pdf |
| 65 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-9 | -2.155e+01 | 0.0000 | 785.0 | 17.97% | 513.4 | 14.60% | motif file (matrix) |
pdf |
| 66 |  | NF1:FOXA1/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-9 | -2.129e+01 | 0.0000 | 57.0 | 1.30% | 18.7 | 0.53% | motif file (matrix) |
pdf |
| 67 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-8 | -1.984e+01 | 0.0000 | 91.0 | 2.08% | 37.4 | 1.06% | motif file (matrix) |
pdf |
| 68 |  | Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer | 1e-8 | -1.954e+01 | 0.0000 | 162.0 | 3.71% | 80.9 | 2.30% | motif file (matrix) |
pdf |
| 69 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-8 | -1.845e+01 | 0.0000 | 467.0 | 10.69% | 290.6 | 8.26% | motif file (matrix) |
pdf |
| 70 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-7 | -1.833e+01 | 0.0000 | 80.0 | 1.83% | 32.9 | 0.94% | motif file (matrix) |
pdf |
| 71 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-7 | -1.831e+01 | 0.0000 | 183.0 | 4.19% | 95.6 | 2.72% | motif file (matrix) |
pdf |
| 72 |  | ETS(ETS)/Promoter/Homer | 1e-7 | -1.802e+01 | 0.0000 | 159.0 | 3.64% | 80.1 | 2.28% | motif file (matrix) |
pdf |
| 73 |  | CArG(MADS)/PUER-Srf-ChIP-Seq(Sullivan et al.)/Homer | 1e-7 | -1.802e+01 | 0.0000 | 222.0 | 5.08% | 121.1 | 3.44% | motif file (matrix) |
pdf |
| 74 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.782e+01 | 0.0000 | 241.0 | 5.52% | 134.4 | 3.82% | motif file (matrix) |
pdf |
| 75 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-7 | -1.751e+01 | 0.0000 | 1675.0 | 38.35% | 1209.0 | 34.38% | motif file (matrix) |
pdf |
| 76 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-7 | -1.717e+01 | 0.0000 | 422.0 | 9.66% | 261.1 | 7.42% | motif file (matrix) |
pdf |
| 77 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-7 | -1.705e+01 | 0.0000 | 942.0 | 21.57% | 645.8 | 18.37% | motif file (matrix) |
pdf |
| 78 |  | AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer | 1e-7 | -1.686e+01 | 0.0000 | 1990.0 | 45.56% | 1461.9 | 41.58% | motif file (matrix) |
pdf |
| 79 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-7 | -1.655e+01 | 0.0000 | 93.0 | 2.13% | 41.2 | 1.17% | motif file (matrix) |
pdf |
| 80 |  | NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-7 | -1.643e+01 | 0.0000 | 1094.0 | 25.05% | 763.5 | 21.71% | motif file (matrix) |
pdf |
| 81 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-7 | -1.625e+01 | 0.0000 | 18.0 | 0.41% | 3.8 | 0.11% | motif file (matrix) |
pdf |
| 82 |  | BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer | 1e-6 | -1.511e+01 | 0.0000 | 981.0 | 22.46% | 682.8 | 19.42% | motif file (matrix) |
pdf |
| 83 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer | 1e-6 | -1.439e+01 | 0.0000 | 304.0 | 6.96% | 184.5 | 5.25% | motif file (matrix) |
pdf |
| 84 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-5 | -1.380e+01 | 0.0000 | 1055.0 | 24.15% | 744.0 | 21.16% | motif file (matrix) |
pdf |
| 85 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-5 | -1.379e+01 | 0.0000 | 336.0 | 7.69% | 208.4 | 5.93% | motif file (matrix) |
pdf |
| 86 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.361e+01 | 0.0000 | 1106.0 | 25.32% | 784.6 | 22.31% | motif file (matrix) |
pdf |
| 87 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-5 | -1.309e+01 | 0.0000 | 693.0 | 15.87% | 472.3 | 13.43% | motif file (matrix) |
pdf |
| 88 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-5 | -1.294e+01 | 0.0000 | 1465.0 | 33.54% | 1066.0 | 30.32% | motif file (matrix) |
pdf |
| 89 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-5 | -1.167e+01 | 0.0000 | 2501.0 | 57.26% | 1899.8 | 54.03% | motif file (matrix) |
pdf |
| 90 |  | ZNF711(Zf)/SH-SY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-4 | -1.102e+01 | 0.0000 | 911.0 | 20.86% | 646.8 | 18.39% | motif file (matrix) |
pdf |
| 91 |  | PAX5-shortForm(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-4 | -1.075e+01 | 0.0001 | 70.0 | 1.60% | 34.0 | 0.97% | motif file (matrix) |
pdf |
| 92 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.059e+01 | 0.0001 | 222.0 | 5.08% | 135.9 | 3.86% | motif file (matrix) |
pdf |
| 93 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-4 | -1.031e+01 | 0.0001 | 269.0 | 6.16% | 169.5 | 4.82% | motif file (matrix) |
pdf |
| 94 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-4 | -9.932e+00 | 0.0001 | 241.0 | 5.52% | 150.9 | 4.29% | motif file (matrix) |
pdf |
| 95 | 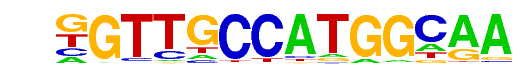 | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-4 | -9.687e+00 | 0.0002 | 125.0 | 2.86% | 70.8 | 2.01% | motif file (matrix) |
pdf |
| 96 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -9.215e+00 | 0.0003 | 815.0 | 18.66% | 581.3 | 16.53% | motif file (matrix) |
pdf |
| 97 |  | LXRE(NR/DR4)/BLRP(RAW)-LXRb-ChIP-Seq(GSE21512)/Homer | 1e-3 | -9.179e+00 | 0.0003 | 26.0 | 0.60% | 9.6 | 0.27% | motif file (matrix) |
pdf |
| 98 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-3 | -8.860e+00 | 0.0004 | 534.0 | 12.23% | 369.4 | 10.50% | motif file (matrix) |
pdf |
| 99 |  | NFAT:AP1/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-3 | -8.773e+00 | 0.0004 | 159.0 | 3.64% | 95.4 | 2.71% | motif file (matrix) |
pdf |
| 100 |  | ETS:E-box/HPC7-Scl-ChIP-Seq(GSE22178)/Homer | 1e-3 | -8.478e+00 | 0.0005 | 61.0 | 1.40% | 30.7 | 0.87% | motif file (matrix) |
pdf |
| 101 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-3 | -7.737e+00 | 0.0011 | 986.0 | 22.57% | 721.2 | 20.51% | motif file (matrix) |
pdf |
| 102 |  | Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-3 | -7.698e+00 | 0.0011 | 75.0 | 1.72% | 40.5 | 1.15% | motif file (matrix) |
pdf |
| 103 |  | RARg(NR)/ES-RARg-ChIP-Seq(GSE30538)/Homer | 1e-3 | -7.607e+00 | 0.0012 | 12.0 | 0.27% | 3.7 | 0.11% | motif file (matrix) |
pdf |
| 104 |  | bZIP:IRF/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-3 | -7.592e+00 | 0.0012 | 340.0 | 7.78% | 229.2 | 6.52% | motif file (matrix) |
pdf |
| 105 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.501e+00 | 0.0013 | 702.0 | 16.07% | 503.9 | 14.33% | motif file (matrix) |
pdf |
| 106 | 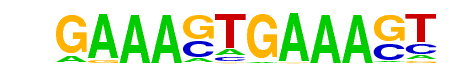 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-2 | -6.753e+00 | 0.0027 | 98.0 | 2.24% | 57.9 | 1.65% | motif file (matrix) |
pdf |
| 107 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -6.669e+00 | 0.0029 | 48.0 | 1.10% | 24.5 | 0.70% | motif file (matrix) |
pdf |
| 108 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -6.614e+00 | 0.0031 | 125.0 | 2.86% | 76.7 | 2.18% | motif file (matrix) |
pdf |
| 109 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-2 | -6.442e+00 | 0.0036 | 1468.0 | 33.61% | 1108.4 | 31.52% | motif file (matrix) |
pdf |
| 110 |  | p63(p53)/Keratinocyte-p63-ChIP-Seq(GSE17611)/Homer | 1e-2 | -6.422e+00 | 0.0036 | 168.0 | 3.85% | 107.1 | 3.05% | motif file (matrix) |
pdf |
| 111 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-2 | -6.065e+00 | 0.0052 | 693.0 | 15.87% | 504.0 | 14.33% | motif file (matrix) |
pdf |
| 112 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-2 | -6.016e+00 | 0.0054 | 59.0 | 1.35% | 32.1 | 0.91% | motif file (matrix) |
pdf |
| 113 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.932e+00 | 0.0058 | 155.0 | 3.55% | 99.9 | 2.84% | motif file (matrix) |
pdf |
| 114 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-2 | -5.717e+00 | 0.0071 | 818.0 | 18.73% | 603.2 | 17.15% | motif file (matrix) |
pdf |
| 115 |  | CRE(bZIP)/Promoter/Homer | 1e-2 | -5.542e+00 | 0.0084 | 120.0 | 2.75% | 75.7 | 2.15% | motif file (matrix) |
pdf |
| 116 |  | EBNA1(EBV virus)/Raji-EBNA1-ChIP-Seq(GSE30709)/Homer | 1e-2 | -5.501e+00 | 0.0087 | 8.0 | 0.18% | 2.7 | 0.08% | motif file (matrix) |
pdf |
| 117 |  | E-box(HLH)/Promoter/Homer | 1e-2 | -5.139e+00 | 0.0123 | 34.0 | 0.78% | 17.2 | 0.49% | motif file (matrix) |
pdf |
| 118 |  | PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer | 1e-2 | -5.026e+00 | 0.0137 | 161.0 | 3.69% | 106.4 | 3.02% | motif file (matrix) |
pdf |
| 119 |  | NFkB-p50,p52(RHD)/p50-ChIP-Chip(Schreiber et al.)/Homer | 1e-2 | -4.968e+00 | 0.0144 | 46.0 | 1.05% | 25.4 | 0.72% | motif file (matrix) |
pdf |
| 120 |  | Reverb(NR/DR2)/BLRP(RAW)-Reverba-ChIP-Seq(GSE45914)/Homer | 1e-2 | -4.962e+00 | 0.0144 | 74.0 | 1.69% | 44.8 | 1.27% | motif file (matrix) |
pdf |
| 121 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.907e+00 | 0.0150 | 283.0 | 6.48% | 197.7 | 5.62% | motif file (matrix) |
pdf |
| 122 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.818e+00 | 0.0163 | 102.0 | 2.34% | 64.7 | 1.84% | motif file (matrix) |
pdf |
| 123 |  | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-2 | -4.753e+00 | 0.0173 | 50.0 | 1.14% | 28.4 | 0.81% | motif file (matrix) |
pdf |
| 124 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-2 | -4.695e+00 | 0.0181 | 1422.0 | 32.55% | 1086.4 | 30.90% | motif file (matrix) |
pdf |
| 125 |  | USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.681e+00 | 0.0182 | 223.0 | 5.11% | 153.1 | 4.35% | motif file (matrix) |
pdf |
| 126 |  | PR(NR)/T47D-PR-ChIP-Seq(GSE31130)/Homer | 1e-2 | -4.624e+00 | 0.0192 | 1343.0 | 30.75% | 1024.4 | 29.13% | motif file (matrix) |
pdf |



























































































































































































































































