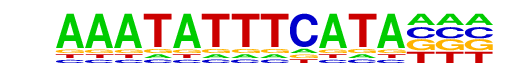
| p-value: | 1e-72 |
| log p-value: | -1.673e+02 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 104.0 |
| Percentage of Target Sequences with motif | 0.38% |
| Number of Background Sequences with motif | 1.8 |
| Percentage of Background Sequences with motif | 0.06% |
| Average Position of motif in Targets | 103.5 +/- 48.8bp |
| Average Position of motif in Background | 143.5 +/- 5.6bp |
| Strand Bias (log2 ratio + to - strand density) | 0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0002.1_Arid5a_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAATATTTCATA-
CTAATATTGCTAAA |
|


|
|
PB0148.1_Mtf1_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.62
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AAATATTTCATA---
-NNTTTTTCTTATNT |
|

|
|
MF0006.1_bZIP_cEBP-like_subclass/Jaspar
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | AAATATTTCATA-
----ATTGCATAA |
|


|
|
PH0172.1_Tlx2/Jaspar
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAATATTTCATA----
TAATTAATTAATAACTA |
|


|
|
MA0466.1_CEBPB/Jaspar
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | AAATATTTCATA--
---TATTGCACAAT |
|


|
|
MF0010.1_Homeobox_class/Jaspar
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AAATATTTCATA
-AATAATT---- |
|


|
|
PB0105.1_Arid3a_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | AAATATTTCATA-----
--NNATNTGATANNNNN |
|


|
|
MA0025.1_NFIL3/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | AAATATTTCATA-
--ANGTTACATAA |
|


|
|
MA0102.3_CEBPA/Jaspar
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | AAATATTTCATA---
----ATTGCACAATA |
|


|
|
PB0096.1_Zfp187_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AAATATTTCATA-
TTATTAGTACATAN |
|


|
|





















