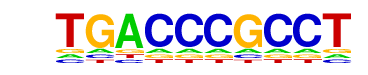
| p-value: | 1e-60 |
| log p-value: | -1.397e+02 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 123.0 |
| Percentage of Target Sequences with motif | 0.45% |
| Number of Background Sequences with motif | 2.3 |
| Percentage of Background Sequences with motif | 0.07% |
| Average Position of motif in Targets | 93.2 +/- 54.7bp |
| Average Position of motif in Background | 98.2 +/- 89.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
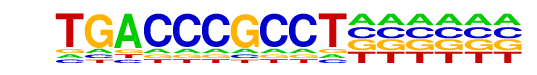
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
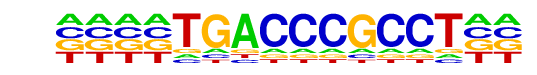
PB0153.1_Nr2f2_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.79
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TGACCCGCCT--
NNNNTGACCCGGCGCG |
|

|
|
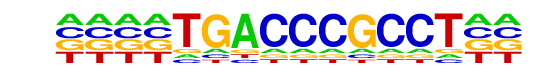
PB0157.1_Rara_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.76
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TGACCCGCCT--
NNCNTGACCCCGCTCT |
|

|
|
PB0180.1_Sp4_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGACCCGCCT---
NNGGCCACGCCTTTN |
|


|
|
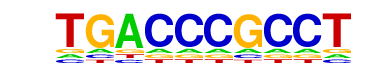
MF0004.1_Nuclear_Receptor_class/Jaspar
| Match Rank: | 4 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGACCCGCCT
TGACCT---- |
|

|
|
MA0516.1_SP2/Jaspar
| Match Rank: | 5 |
| Score: | 0.70
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TGACCCGCCT------
-GCCCCGCCCCCTCCC |
|

|
|
PB0164.1_Smad3_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TGACCCGCCT-----
TACGCCCCGCCACTCTG |
|


|
|
PB0025.1_Glis2_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.66
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TGACCCGCCT---
TATCGACCCCCCACAG |
|


|
|
PB0024.1_Gcm1_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TGACCCGCCT----
TCGTACCCGCATCATT |
|


|
|
Sp1(Zf)/Promoter/Homer
| Match Rank: | 9 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TGACCCGCCT--
GGCCCCGCCCCC |
|


|
|
MA0079.3_SP1/Jaspar
| Match Rank: | 10 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TGACCCGCCT--
-GCCCCGCCCCC |
|


|
|