| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | E-box(HLH)/Promoter/Homer | 1e-20 | -4.625e+01 | 0.0000 | 102.0 | 0.38% | 5.0 | 0.16% | motif file (matrix) |
pdf |
| 2 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-16 | -3.802e+01 | 0.0000 | 326.0 | 1.21% | 23.2 | 0.74% | motif file (matrix) |
pdf |
| 3 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-16 | -3.747e+01 | 0.0000 | 336.0 | 1.24% | 24.1 | 0.76% | motif file (matrix) |
pdf |
| 4 |  | HRE(HSF)/HepG2-HSF1-ChIP-Seq(GSE31477)/Homer | 1e-14 | -3.258e+01 | 0.0000 | 369.0 | 1.36% | 28.7 | 0.91% | motif file (matrix) |
pdf |
| 5 |  | CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-12 | -2.819e+01 | 0.0000 | 1101.0 | 4.07% | 104.0 | 3.30% | motif file (matrix) |
pdf |
| 6 |  | E2F(E2F)/Cell-Cycle-Exp/Homer | 1e-11 | -2.664e+01 | 0.0000 | 36.0 | 0.13% | 1.1 | 0.03% | motif file (matrix) |
pdf |
| 7 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-11 | -2.606e+01 | 0.0000 | 1063.0 | 3.93% | 100.2 | 3.18% | motif file (matrix) |
pdf |
| 8 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-11 | -2.572e+01 | 0.0000 | 156.0 | 0.58% | 10.2 | 0.32% | motif file (matrix) |
pdf |
| 9 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-10 | -2.506e+01 | 0.0000 | 504.0 | 1.86% | 43.7 | 1.39% | motif file (matrix) |
pdf |
| 10 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-10 | -2.319e+01 | 0.0000 | 2665.0 | 9.85% | 275.6 | 8.75% | motif file (matrix) |
pdf |
| 11 |  | Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer | 1e-9 | -2.140e+01 | 0.0000 | 1079.0 | 3.99% | 105.0 | 3.33% | motif file (matrix) |
pdf |
| 12 |  | STAT6/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-9 | -2.135e+01 | 0.0000 | 1536.0 | 5.68% | 153.7 | 4.88% | motif file (matrix) |
pdf |
| 13 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-9 | -2.122e+01 | 0.0000 | 76.0 | 0.28% | 4.8 | 0.15% | motif file (matrix) |
pdf |
| 14 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-8 | -2.003e+01 | 0.0000 | 244.0 | 0.90% | 19.2 | 0.61% | motif file (matrix) |
pdf |
| 15 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-8 | -1.969e+01 | 0.0000 | 777.0 | 2.87% | 73.5 | 2.34% | motif file (matrix) |
pdf |
| 16 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-8 | -1.953e+01 | 0.0000 | 2102.0 | 7.77% | 216.8 | 6.89% | motif file (matrix) |
pdf |
| 17 |  | ZNF711(Zf)/SH-SY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-7 | -1.842e+01 | 0.0000 | 3373.0 | 12.47% | 358.3 | 11.38% | motif file (matrix) |
pdf |
| 18 |  | EBNA1(EBV virus)/Raji-EBNA1-ChIP-Seq(GSE30709)/Homer | 1e-7 | -1.842e+01 | 0.0000 | 30.0 | 0.11% | 1.7 | 0.06% | motif file (matrix) |
pdf |
| 19 | 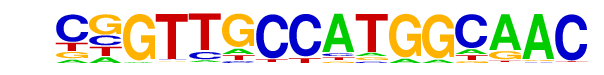 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-7 | -1.800e+01 | 0.0000 | 218.0 | 0.81% | 17.9 | 0.57% | motif file (matrix) |
pdf |
| 20 |  | c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer | 1e-7 | -1.778e+01 | 0.0000 | 673.0 | 2.49% | 63.2 | 2.01% | motif file (matrix) |
pdf |
| 21 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-7 | -1.770e+01 | 0.0000 | 5583.0 | 20.64% | 608.1 | 19.31% | motif file (matrix) |
pdf |
| 22 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-7 | -1.738e+01 | 0.0000 | 8199.0 | 30.32% | 907.0 | 28.81% | motif file (matrix) |
pdf |
| 23 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-7 | -1.726e+01 | 0.0000 | 536.0 | 1.98% | 49.7 | 1.58% | motif file (matrix) |
pdf |
| 24 |  | GLI3(Zf)/GLI3-ChIP-Chip(GSE11077)/Homer | 1e-7 | -1.699e+01 | 0.0000 | 226.0 | 0.84% | 18.3 | 0.58% | motif file (matrix) |
pdf |
| 25 |  | Pax7-longest(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-6 | -1.586e+01 | 0.0000 | 69.0 | 0.26% | 4.8 | 0.15% | motif file (matrix) |
pdf |
| 26 |  | HRE(HSF)/Striatum-HSF1-ChIP-Seq(GSE38000)/Homer | 1e-6 | -1.527e+01 | 0.0000 | 595.0 | 2.20% | 56.8 | 1.80% | motif file (matrix) |
pdf |
| 27 |  | ETS:E-box/HPC7-Scl-ChIP-Seq(GSE22178)/Homer | 1e-6 | -1.471e+01 | 0.0000 | 220.0 | 0.81% | 18.5 | 0.59% | motif file (matrix) |
pdf |
| 28 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-6 | -1.463e+01 | 0.0000 | 3913.0 | 14.47% | 423.0 | 13.44% | motif file (matrix) |
pdf |
| 29 |  | NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-6 | -1.409e+01 | 0.0000 | 2419.0 | 8.94% | 256.9 | 8.16% | motif file (matrix) |
pdf |
| 30 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-5 | -1.339e+01 | 0.0000 | 6169.0 | 22.81% | 682.0 | 21.66% | motif file (matrix) |
pdf |
| 31 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-5 | -1.333e+01 | 0.0000 | 175.0 | 0.65% | 14.2 | 0.45% | motif file (matrix) |
pdf |
| 32 |  | HOXD13(Homeobox)/Chicken-Hoxd13-ChIP-Seq(GSE38910)/Homer | 1e-5 | -1.331e+01 | 0.0000 | 3117.0 | 11.53% | 335.4 | 10.65% | motif file (matrix) |
pdf |
| 33 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-5 | -1.305e+01 | 0.0000 | 1671.0 | 6.18% | 174.3 | 5.54% | motif file (matrix) |
pdf |
| 34 |  | Tbox:Smad/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer | 1e-5 | -1.292e+01 | 0.0000 | 489.0 | 1.81% | 46.7 | 1.48% | motif file (matrix) |
pdf |
| 35 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-5 | -1.245e+01 | 0.0000 | 4329.0 | 16.01% | 473.3 | 15.03% | motif file (matrix) |
pdf |
| 36 |  | Hoxc9(Homeobox)/Ainv15-Hoxc9-ChIP-Seq(GSE21812)/Homer | 1e-5 | -1.232e+01 | 0.0000 | 1393.0 | 5.15% | 144.7 | 4.60% | motif file (matrix) |
pdf |
| 37 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-5 | -1.204e+01 | 0.0000 | 1762.0 | 6.52% | 185.5 | 5.89% | motif file (matrix) |
pdf |
| 38 |  | Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer | 1e-5 | -1.183e+01 | 0.0000 | 635.0 | 2.35% | 63.0 | 2.00% | motif file (matrix) |
pdf |
| 39 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-5 | -1.179e+01 | 0.0000 | 3950.0 | 14.61% | 431.5 | 13.71% | motif file (matrix) |
pdf |
| 40 |  | Unknown(Homeobox)/Limb-p300-ChIP-Seq/Homer | 1e-5 | -1.171e+01 | 0.0001 | 1741.0 | 6.44% | 183.0 | 5.81% | motif file (matrix) |
pdf |
| 41 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-5 | -1.163e+01 | 0.0001 | 783.0 | 2.90% | 78.3 | 2.49% | motif file (matrix) |
pdf |
| 42 |  | E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -1.139e+01 | 0.0001 | 2998.0 | 11.09% | 324.9 | 10.32% | motif file (matrix) |
pdf |
| 43 |  | BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer | 1e-4 | -1.063e+01 | 0.0001 | 3926.0 | 14.52% | 430.1 | 13.66% | motif file (matrix) |
pdf |
| 44 |  | Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-4 | -1.057e+01 | 0.0001 | 61.0 | 0.23% | 4.7 | 0.15% | motif file (matrix) |
pdf |
| 45 |  | NFY(CCAAT)/Promoter/Homer | 1e-4 | -1.048e+01 | 0.0002 | 1640.0 | 6.06% | 173.8 | 5.52% | motif file (matrix) |
pdf |
| 46 |  | PRDM9(Zf)/Testis-DMC1-ChIP-Seq(GSE35498)/Homer | 1e-4 | -1.026e+01 | 0.0002 | 940.0 | 3.48% | 96.5 | 3.06% | motif file (matrix) |
pdf |
| 47 |  | CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-4 | -1.017e+01 | 0.0002 | 343.0 | 1.27% | 32.4 | 1.03% | motif file (matrix) |
pdf |
| 48 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-3 | -8.869e+00 | 0.0007 | 866.0 | 3.20% | 89.5 | 2.84% | motif file (matrix) |
pdf |
| 49 |  | NFAT:AP1/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-3 | -8.598e+00 | 0.0009 | 421.0 | 1.56% | 41.7 | 1.32% | motif file (matrix) |
pdf |
| 50 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-3 | -8.571e+00 | 0.0009 | 3175.0 | 11.74% | 348.1 | 11.05% | motif file (matrix) |
pdf |
| 51 |  | TATA-Box(TBP)/Promoter/Homer | 1e-3 | -8.441e+00 | 0.0010 | 3807.0 | 14.08% | 420.3 | 13.35% | motif file (matrix) |
pdf |
| 52 |  | CHR/Cell-Cycle-Exp/Homer | 1e-3 | -8.404e+00 | 0.0011 | 1763.0 | 6.52% | 190.0 | 6.03% | motif file (matrix) |
pdf |
| 53 | 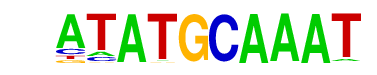 | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.050e+00 | 0.0015 | 723.0 | 2.67% | 74.2 | 2.36% | motif file (matrix) |
pdf |
| 54 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-3 | -8.048e+00 | 0.0015 | 2585.0 | 9.56% | 282.2 | 8.96% | motif file (matrix) |
pdf |
| 55 |  | Nkx2.5(Homeobox)/HL1-Nkx2.5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-3 | -7.983e+00 | 0.0015 | 5777.0 | 21.36% | 646.9 | 20.55% | motif file (matrix) |
pdf |
| 56 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-3 | -7.792e+00 | 0.0018 | 1030.0 | 3.81% | 108.6 | 3.45% | motif file (matrix) |
pdf |
| 57 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-3 | -7.467e+00 | 0.0025 | 2427.0 | 8.97% | 266.0 | 8.45% | motif file (matrix) |
pdf |
| 58 |  | Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer | 1e-3 | -7.214e+00 | 0.0031 | 7371.0 | 27.25% | 831.2 | 26.40% | motif file (matrix) |
pdf |
| 59 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.132e+00 | 0.0033 | 625.0 | 2.31% | 64.9 | 2.06% | motif file (matrix) |
pdf |
| 60 |  | CRE(bZIP)/Promoter/Homer | 1e-3 | -7.123e+00 | 0.0033 | 357.0 | 1.32% | 35.6 | 1.13% | motif file (matrix) |
pdf |
| 61 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.067e+00 | 0.0034 | 734.0 | 2.71% | 76.9 | 2.44% | motif file (matrix) |
pdf |
| 62 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-3 | -6.981e+00 | 0.0037 | 166.0 | 0.61% | 15.4 | 0.49% | motif file (matrix) |
pdf |
| 63 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-2 | -6.652e+00 | 0.0050 | 1592.0 | 5.89% | 172.4 | 5.47% | motif file (matrix) |
pdf |
| 64 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-2 | -6.635e+00 | 0.0050 | 7948.0 | 29.39% | 899.3 | 28.56% | motif file (matrix) |
pdf |
| 65 |  | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-2 | -6.630e+00 | 0.0050 | 3883.0 | 14.36% | 432.1 | 13.72% | motif file (matrix) |
pdf |
| 66 |  | PAX5(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.587e+00 | 0.0051 | 703.0 | 2.60% | 73.8 | 2.34% | motif file (matrix) |
pdf |
| 67 |  | bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.577e+00 | 0.0051 | 502.0 | 1.86% | 51.2 | 1.63% | motif file (matrix) |
pdf |
| 68 |  | NFAT(RHD)/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-2 | -6.219e+00 | 0.0072 | 2295.0 | 8.49% | 252.2 | 8.01% | motif file (matrix) |
pdf |
| 69 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-2 | -6.110e+00 | 0.0079 | 1763.0 | 6.52% | 193.0 | 6.13% | motif file (matrix) |
pdf |
| 70 |  | HNF4a(NR/DR1)/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-2 | -6.085e+00 | 0.0080 | 1095.0 | 4.05% | 117.1 | 3.72% | motif file (matrix) |
pdf |
| 71 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-2 | -6.080e+00 | 0.0080 | 220.0 | 0.81% | 21.2 | 0.67% | motif file (matrix) |
pdf |
| 72 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-2 | -5.940e+00 | 0.0090 | 3249.0 | 12.01% | 362.0 | 11.50% | motif file (matrix) |
pdf |
| 73 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-2 | -5.740e+00 | 0.0108 | 4417.0 | 16.33% | 495.7 | 15.75% | motif file (matrix) |
pdf |
| 74 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-2 | -5.698e+00 | 0.0111 | 5818.0 | 21.51% | 656.1 | 20.84% | motif file (matrix) |
pdf |
| 75 |  | Tlx?(NR)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-2 | -5.418e+00 | 0.0145 | 936.0 | 3.46% | 100.2 | 3.18% | motif file (matrix) |
pdf |
| 76 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.292e+00 | 0.0163 | 4216.0 | 15.59% | 473.7 | 15.05% | motif file (matrix) |
pdf |
| 77 |  | VDR(NR/DR3)/GM10855-VDR+vitD-ChIP-Seq(GSE22484)/Homer | 1e-2 | -5.251e+00 | 0.0167 | 383.0 | 1.42% | 39.0 | 1.24% | motif file (matrix) |
pdf |
| 78 |  | ETS(ETS)/Promoter/Homer | 1e-2 | -5.226e+00 | 0.0170 | 547.0 | 2.02% | 57.3 | 1.82% | motif file (matrix) |
pdf |
| 79 |  | Mouse_Recombination_Hotspot/Testis-DMC1-ChIP-Seq(GSE24438)/Homer | 1e-2 | -5.141e+00 | 0.0182 | 130.0 | 0.48% | 12.4 | 0.39% | motif file (matrix) |
pdf |
| 80 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.085e+00 | 0.0190 | 187.0 | 0.69% | 18.4 | 0.58% | motif file (matrix) |
pdf |
| 81 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.047e+00 | 0.0195 | 2286.0 | 8.45% | 253.4 | 8.05% | motif file (matrix) |
pdf |
| 82 |  | STAT6(Stat)/CD4-Stat6-ChIP-Seq(GSE22104)/Homer | 1e-2 | -4.962e+00 | 0.0210 | 1500.0 | 5.55% | 164.4 | 5.22% | motif file (matrix) |
pdf |
| 83 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-2 | -4.935e+00 | 0.0213 | 644.0 | 2.38% | 68.2 | 2.17% | motif file (matrix) |
pdf |
| 84 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-2 | -4.649e+00 | 0.0277 | 213.0 | 0.79% | 21.0 | 0.67% | motif file (matrix) |
pdf |
| 85 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-2 | -4.649e+00 | 0.0277 | 213.0 | 0.79% | 21.0 | 0.67% | motif file (matrix) |
pdf |









































































































































































