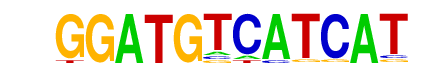
| p-value: | 1e-12 |
| log p-value: | -2.959e+01 |
| Information Content per bp: | 1.863 |
| Number of Target Sequences with motif | 12.0 |
| Percentage of Target Sequences with motif | 0.38% |
| Number of Background Sequences with motif | 4.4 |
| Percentage of Background Sequences with motif | 0.02% |
| Average Position of motif in Targets | 94.8 +/- 58.0bp |
| Average Position of motif in Background | 135.1 +/- 20.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.5 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0488.1_JUN/Jaspar
| Match Rank: | 1 |
| Score: | 0.78
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GGATGTCATCAT---
--ATGACATCATCNN |
|


|
|
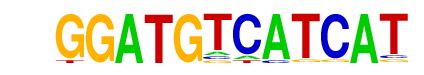
c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 2 |
| Score: | 0.75
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GGATGTCATCAT
NGATGACGTCAT |
|

|
|
MA0492.1_JUND_(var.2)/Jaspar
| Match Rank: | 3 |
| Score: | 0.75
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GGATGTCATCAT----
-NATGACATCATCNNN |
|


|
|
PH0014.1_Cphx/Jaspar
| Match Rank: | 4 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GGATGTCATCAT
NTTGATTNNATCAN |
|


|
|
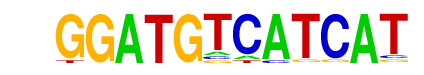
JunD(bZIP)/K562-JunD-ChIP-Seq/Homer
| Match Rank: | 5 |
| Score: | 0.72
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GGATGTCATCAT
NGATGACGTCAT |
|

|
|
GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer
| Match Rank: | 6 |
| Score: | 0.68
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------GGATGTCATCAT--
NNNNNBAGATAWYATCTVHN |
|


|
|
PB0038.1_Jundm2_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGATGTCATCAT---
CCGATGACGTCATCGT |
|


|
|
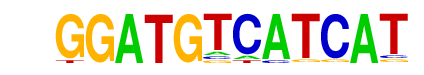
Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 8 |
| Score: | 0.66
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GGATGTCATCAT
--ATTGCATCAT |
|

|
|
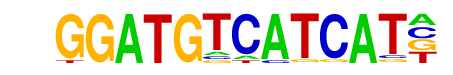
Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer
| Match Rank: | 9 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GGATGTCATCAT-
-SCTGTCARCACC |
|

|
|
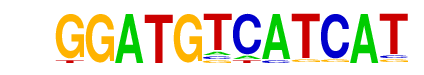
Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 10 |
| Score: | 0.65
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GGATGTCATCAT
--ATTGCATCAK |
|

|
|