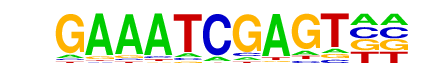
| p-value: | 1e-13 |
| log p-value: | -3.022e+01 |
| Information Content per bp: | 1.651 |
| Number of Target Sequences with motif | 80.0 |
| Percentage of Target Sequences with motif | 2.54% |
| Number of Background Sequences with motif | 264.4 |
| Percentage of Background Sequences with motif | 0.98% |
| Average Position of motif in Targets | 105.6 +/- 58.3bp |
| Average Position of motif in Background | 106.1 +/- 54.9bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PH0044.1_Homez/Jaspar
| Match Rank: | 1 |
| Score: | 0.62
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GAAATCGAGT----
NNTAAAAACGATGTTNT |
|


|
|
PH0037.1_Hdx/Jaspar
| Match Rank: | 2 |
| Score: | 0.62
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GAAATCGAGT--
AAGGCGAAATCATCGCA |
|


|
|
IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 3 |
| Score: | 0.61
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GAAATCGAGT
ACTGAAACCA--- |
|


|
|
Pax7(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer
| Match Rank: | 4 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GAAATCGAGT-
-TAATCAATTA |
|


|
|
EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----GAAATCGAGT
NACAGGAAAT----- |
|


|
|
MA0156.1_FEV/Jaspar
| Match Rank: | 6 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GAAATCGAGT
CAGGAAAT----- |
|


|
|
PB0139.1_Irf5_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GAAATCGAGT-----
TTGACCGAGAATTCC |
|


|
|
ISRE(IRF)/ThioMac-LPS-exp(GSE23622)/HOMER
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GAAATCGAGT--
GAAACTGAAACT |
|

|
|
HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer
| Match Rank: | 9 |
| Score: | 0.55
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GAAATCGAGT-
-DGATCRATAN |
|


|
|
MA0051.1_IRF2/Jaspar
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GAAATCGAGT-------
GGAAAGCGAAACCAAAAC |
|


|
|





















