| p-value: | 1e-45 |
| log p-value: | -1.039e+02 |
| Information Content per bp: | 1.694 |
| Number of Target Sequences with motif | 58.0 |
| Percentage of Target Sequences with motif | 0.34% |
| Number of Background Sequences with motif | 3.7 |
| Percentage of Background Sequences with motif | 0.03% |
| Average Position of motif in Targets | 88.9 +/- 49.1bp |
| Average Position of motif in Background | 132.0 +/- 28.4bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PH0148.1_Pou3f3/Jaspar
| Match Rank: | 1 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | reverse strand |
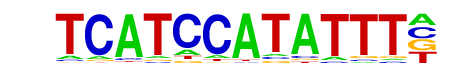
| Alignment: | ----TCATCCATATTT-
TNNATTATGCATANNTT |
|


|
|
HOXA2(Homeobox)/mES-Hoxa2-ChIP-Seq(Donaldson et al.)/Homer
| Match Rank: | 2 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TCATCCATATTT
GYCATCMATCAT- |
|


|
|
MA0095.2_YY1/Jaspar
| Match Rank: | 3 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TCATCCATATTT
GCNGCCATCTTG |
|


|
|
PB0028.1_Hbp1_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TCATCCATATTT
NNCATTCATTCATNNN- |
|


|
|
PB0002.1_Arid5a_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TCATCCATATTT-
NNTNNCAATATTAG |
|


|
|
MF0008.1_MADS_class/Jaspar
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | TCATCCATATTT--
----CCATATATGG |
|


|
|
MA0507.1_POU2F2/Jaspar
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TCATCCATATTT-
ATATGCAAATNNN |
|

|
|
PB0146.1_Mafk_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TCATCCATATTT---
CCTTGCAATTTTTNN |
|


|
|
PB0078.1_Srf_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | TCATCCATATTT----
--TTCCATATATGGAA |
|


|
|
PH0126.1_Obox6/Jaspar
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TCATCCATATTT
CNATAATCCGNTTNT |
|


|
|





















