| p-value: | 1e-44 |
| log p-value: | -1.035e+02 |
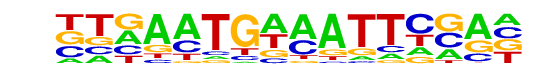
| Information Content per bp: | 1.840 |
| Number of Target Sequences with motif | 41.0 |
| Percentage of Target Sequences with motif | 0.24% |
| Number of Background Sequences with motif | 1.7 |
| Percentage of Background Sequences with motif | 0.01% |
| Average Position of motif in Targets | 108.5 +/- 50.7bp |
| Average Position of motif in Background | 123.3 +/- 54.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer
| Match Rank: | 1 |
| Score: | 0.60
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATCAGATTCAAG
TAATCHGATTAC-- |
|


|
|
PB0170.1_Sox17_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ATCAGATTCAAG----
GACCACATTCATACAAT |
|


|
|
Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer
| Match Rank: | 3 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATCAGATTCAAG
GKVTCADRTTWC-- |
|


|
|
PB0105.1_Arid3a_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.57
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------ATCAGATTCAAG
ACCCGTATCAAATTT--- |
|


|
|
PB0169.1_Sox15_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.56
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----ATCAGATTCAAG
TTGAATGAAATTCGA- |
|

|
|
Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer
| Match Rank: | 6 |
| Score: | 0.55
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --ATCAGATTCAAG
TAATYNRATTAR-- |
|


|
|
PB0176.1_Sox5_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ATCAGATTCAAG--
TATCATAATTAAGGA |
|


|
|
CHR/Cell-Cycle-Exp/Homer
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | ATCAGATTCAAG
--CGGTTTCAAA |
|


|
|
PB0178.1_Sox8_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | ATCAGATTCAAG-----
---ACATTCATGACACG |
|


|
|
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | ATCAGATTCAAG-
---GCATTCCAGN |
|


|
|





















