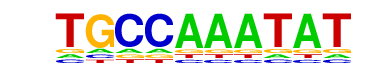
| p-value: | 1e-84 |
| log p-value: | -1.950e+02 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 317.0 |
| Percentage of Target Sequences with motif | 16.34% |
| Number of Background Sequences with motif | 4.8 |
| Percentage of Background Sequences with motif | 5.40% |
| Average Position of motif in Targets | 100.5 +/- 55.2bp |
| Average Position of motif in Background | 125.7 +/- 57.2bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.12 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0161.1_NFIC/Jaspar
| Match Rank: | 1 |
| Score: | 0.73
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCCAAATAT
TGCCAA---- |
|

|
|
NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGCCAAATAT
TTGCCAAG--- |
|


|
|
MF0008.1_MADS_class/Jaspar
| Match Rank: | 3 |
| Score: | 0.69
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TGCCAAATAT--
--CCATATATGG |
|


|
|
PB0078.1_Srf_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCCAAATAT----
TNCCATATATGGNA |
|


|
|
CArG(MADS)/PUER-Srf-ChIP-Seq(Sullivan et al.)/Homer
| Match Rank: | 5 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCCAAATAT--
TNCCATATATGG |
|


|
|
PB0002.1_Arid5a_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.63
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | TGCCAAATAT-------
---CTAATATTGCTAAA |
|


|
|
MA0083.2_SRF/Jaspar
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TGCCAAATAT-----
CATGCCCAAATAAGGCAA |
|

|
|
PB0145.1_Mafb_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.61
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----TGCCAAATAT-
CAATTGCAAAAATAT |
|


|
|
PB0133.1_Hic1_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.60
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TGCCAAATAT-
GGGTGTGCCCAAAAGG |
|

|
|
MA0052.2_MEF2A/Jaspar
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TGCCAAATAT-----
AGCTAAAAATAGCAT |
|


|
|