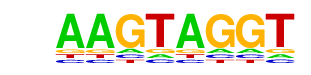
| p-value: | 1e-48 |
| log p-value: | -1.115e+02 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 252.0 |
| Percentage of Target Sequences with motif | 12.99% |
| Number of Background Sequences with motif | 4.0 |
| Percentage of Background Sequences with motif | 4.52% |
| Average Position of motif in Targets | 99.5 +/- 53.6bp |
| Average Position of motif in Background | 143.6 +/- 15.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.07 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
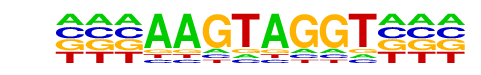
MA0072.1_RORA_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.77
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AAGTAGGT---
TATAAGTAGGTCAA |
|

|
|
MA0032.1_FOXC1/Jaspar
| Match Rank: | 2 |
| Score: | 0.67
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AAGTAGGT
GGTAAGTA--- |
|


|
|
MA0124.1_NKX3-1/Jaspar
| Match Rank: | 3 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AAGTAGGT
TAAGTAT-- |
|


|
|
PH0115.1_Nkx2-6/Jaspar
| Match Rank: | 4 |
| Score: | 0.64
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------AAGTAGGT--
AATNTTAAGTGGNTNN |
|


|
|
PB0048.1_Nkx3-1_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.64
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------AAGTAGGT---
NTNNTTAAGTGGNTNAN |
|


|
|
MA0122.1_Nkx3-2/Jaspar
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AAGTAGGT
TTAAGTGGA- |
|


|
|
SD0001.1_at_AC_acceptor/Jaspar
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAGTAGGT--
CAGGTAAGTAT |
|


|
|
PH0171.1_Nkx2-1/Jaspar
| Match Rank: | 8 |
| Score: | 0.63
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------AAGTAGGT--
AANTTCAAGTGGCTTN |
|


|
|
PH0113.1_Nkx2-4/Jaspar
| Match Rank: | 9 |
| Score: | 0.63
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------AAGTAGGT--
AATTTCAAGTGGCTTN |
|


|
|
PB0155.1_Osr2_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----AAGTAGGT---
NNTGTAGGTAGCANNT |
|


|
|