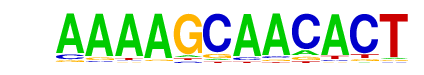
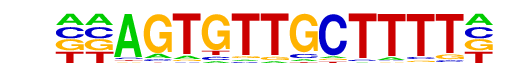
| p-value: | 1e-16 |
| log p-value: | -3.858e+01 |
| Information Content per bp: | 1.699 |
| Number of Target Sequences with motif | 110.0 |
| Percentage of Target Sequences with motif | 5.67% |
| Number of Background Sequences with motif | 2.5 |
| Percentage of Background Sequences with motif | 2.79% |
| Average Position of motif in Targets | 100.3 +/- 56.8bp |
| Average Position of motif in Background | 79.7 +/- 0.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
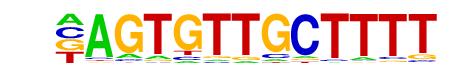
MA0041.1_Foxd3/Jaspar
| Match Rank: | 1 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGTGTTGCTTTT
GAATGTTTGTTT- |
|

|
|
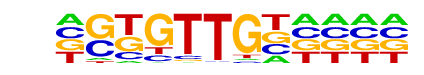
MA0133.1_BRCA1/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AGTGTTGCTTTT
-GTGTTGN---- |
|

|
|
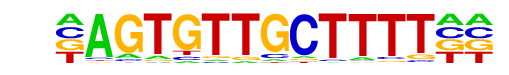
PB0122.1_Foxk1_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGTGTTGCTTTT--
NNNTGTTGTTGTTNG |
|

|
|
IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGTGTTGCTTTT
RSTTTCRSTTTC |
|


|
|
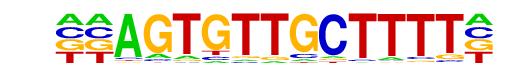
PB0093.1_Zfp105_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AGTGTTGCTTTT-
NTNTTGTTGTTTGTN |
|

|
|
PB0037.1_Isgf3g_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AGTGTTGCTTTT-
TNAGTTTCGATTTTN |
|

|
|
PB0141.1_Isgf3g_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGTGTTGCTTTT-
NNGTANTGTTTTNC |
|


|
|
ISRE(IRF)/ThioMac-LPS-exp(GSE23622)/HOMER
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGTGTTGCTTTT
AGTTTCAGTTTC |
|


|
|
PB0123.1_Foxl1_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGTGTTGCTTTT----
NNTTTTGTTTTGATNT |
|


|
|
Mouse_Recombination_Hotspot/Testis-DMC1-ChIP-Seq(GSE24438)/Homer
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | -9
| | Orientation: | forward strand |
| Alignment: | ---------AGTGTTGCTTTT
ACTYKNATTCGTGNTACTTC- |
|


|
|