| p-value: | 1e-160 |
| log p-value: | -3.705e+02 |
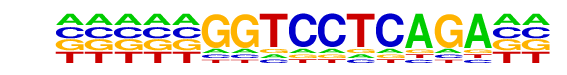
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 374.0 |
| Percentage of Target Sequences with motif | 19.28% |
| Number of Background Sequences with motif | 3.9 |
| Percentage of Background Sequences with motif | 4.44% |
| Average Position of motif in Targets | 96.8 +/- 56.5bp |
| Average Position of motif in Background | 84.6 +/- 49.3bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.11 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
STAT6/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer
| Match Rank: | 1 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GGTCCTCAGA-
-TTCCKNAGAA |
|


|
|
MA0520.1_Stat6/Jaspar
| Match Rank: | 2 |
| Score: | 0.59
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GGTCCTCAGA---
CATTTCCTGAGAAAT |
|


|
|
MA0056.1_MZF1_1-4/Jaspar
| Match Rank: | 3 |
| Score: | 0.57
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GGTCCTCAGA
--TCCCCA-- |
|


|
|
STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer
| Match Rank: | 4 |
| Score: | 0.56
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GGTCCTCAGA---
-TTTCTNAGAAAN |
|


|
|
Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 5 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGTCCTCAGA
CNGTCCTCCC- |
|


|
|
PB0196.1_Zbtb7b_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.53
| | Offset: | -8
| | Orientation: | reverse strand |
| Alignment: | --------GGTCCTCAGA
NNANTGGTGGTCTTNNN- |
|


|
|
MA0519.1_Stat5a::Stat5b/Jaspar
| Match Rank: | 7 |
| Score: | 0.53
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTCCTCAGA-
ATTTCCAAGAA |
|


|
|
MF0004.1_Nuclear_Receptor_class/Jaspar
| Match Rank: | 8 |
| Score: | 0.53
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGTCCTCAGA
AGGTCA----- |
|


|
|
TRa(NR)/C17.2-TRa-ChIP-Seq(GSE38347)/Homer
| Match Rank: | 9 |
| Score: | 0.53
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GGTCCTCAGA-----
TGWCCTCARNTGACC |
|


|
|
PB0099.1_Zfp691_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GGTCCTCAGA--
CGAACAGTGCTCACTAT |
|

|
|





















