| p-value: | 1e-28 |
| log p-value: | -6.595e+01 |
| Information Content per bp: | 1.598 |
| Number of Target Sequences with motif | 37.0 |
| Percentage of Target Sequences with motif | 2.50% |
| Number of Background Sequences with motif | 2.7 |
| Percentage of Background Sequences with motif | 0.25% |
| Average Position of motif in Targets | 94.7 +/- 54.0bp |
| Average Position of motif in Background | 60.9 +/- 42.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0098.1_Zfp410_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TTGTGATGGGAC------
-TATTATGGGATGGATAA |
|


|
|
PH0134.1_Pbx1/Jaspar
| Match Rank: | 2 |
| Score: | 0.58
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTGTGATGGGAC-
NNNNNATTGATGNGTGN |
|


|
|
Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 3 |
| Score: | 0.58
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTGTGATGGGAC
--ATGATGCAAT |
|


|
|
Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 4 |
| Score: | 0.57
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | TTGTGATGGGAC
--MTGATGCAAT |
|


|
|
MA0142.1_Pou5f1::Sox2/Jaspar
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TTGTGATGGGAC-
CTTTGTTATGCAAAT |
|


|
|
OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 6 |
| Score: | 0.56
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTGTGATGGGAC-
CATTGTTATGCAAAT |
|


|
|
RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTGTGATGGGAC
NNHTGTGGTTWN-- |
|


|
|
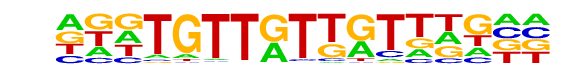
PB0122.1_Foxk1_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TTGTGATGGGAC
NNNTGTTGTTGTTNG-- |
|

|
|
PB0121.1_Foxj3_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTGTGATGGGAC-
NNCTTTGTTTTGNTNNN |
|


|
|
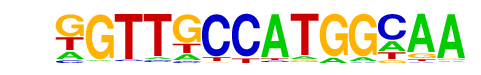
X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TTGTGATGGGAC
GGTTGCCATGGCAA |
|

|
|





















