| p-value: | 1e-18 |
| log p-value: | -4.319e+01 |
| Information Content per bp: | 1.859 |
| Number of Target Sequences with motif | 50.0 |
| Percentage of Target Sequences with motif | 4.43% |
| Number of Background Sequences with motif | 13.8 |
| Percentage of Background Sequences with motif | 0.95% |
| Average Position of motif in Targets | 101.1 +/- 52.9bp |
| Average Position of motif in Background | 91.5 +/- 66.4bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0181.1_Spdef_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.76
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GCATCCKA----
GATAACATCCTAGTAG |
|


|
|
PB0098.1_Zfp410_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GCATCCKA-----
NNNTCCATCCCATAANN |
|


|
|
Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCATCCKA-
NRYTTCCGGH |
|


|
|
PB0077.1_Spdef_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GCATCCKA------
GTACATCCGGATTTTT |
|


|
|
SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 5 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCATCCKA--
ACATCCTGNT |
|


|
|
MA0136.1_ELF5/Jaspar
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCATCCKA
TACTTCCTT |
|


|
|
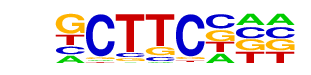
POL008.1_DCE_S_I/Jaspar
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCATCCKA
GCTTCC-- |
|

|
|
MF0001.1_ETS_class/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GCATCCKA-
-CTTCCGGT |
|


|
|
Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCATCCKA-
NRYTTCCGGY |
|


|
|
ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GCATCCKA--
ACTTCCGGTT |
|


|
|





















