| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Reverb(NR/DR2)/BLRP(RAW)-Reverba-ChIP-Seq(GSE45914)/Homer | 1e-7 | -1.786e+01 | 0.0000 | 23.0 | 2.04% | 7.3 | 0.50% | motif file (matrix) |
pdf |
| 2 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-7 | -1.660e+01 | 0.0000 | 93.0 | 8.24% | 66.5 | 4.60% | motif file (matrix) |
pdf |
| 3 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-6 | -1.405e+01 | 0.0001 | 99.0 | 8.78% | 76.8 | 5.32% | motif file (matrix) |
pdf |
| 4 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-5 | -1.352e+01 | 0.0001 | 88.0 | 7.80% | 66.7 | 4.62% | motif file (matrix) |
pdf |
| 5 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-5 | -1.154e+01 | 0.0005 | 106.0 | 9.40% | 88.5 | 6.13% | motif file (matrix) |
pdf |
| 6 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-4 | -1.005e+01 | 0.0018 | 149.0 | 13.21% | 138.4 | 9.58% | motif file (matrix) |
pdf |
| 7 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.279e+00 | 0.0033 | 35.0 | 3.10% | 22.9 | 1.59% | motif file (matrix) |
pdf |
| 8 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-4 | -9.219e+00 | 0.0033 | 28.0 | 2.48% | 16.2 | 1.12% | motif file (matrix) |
pdf |
| 9 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.646e+00 | 0.0048 | 75.0 | 6.65% | 62.9 | 4.35% | motif file (matrix) |
pdf |
| 10 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.210e+00 | 0.0494 | 22.0 | 1.95% | 14.7 | 1.02% | motif file (matrix) |
pdf |
| 11 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-2 | -6.078e+00 | 0.0513 | 84.0 | 7.45% | 78.3 | 5.42% | motif file (matrix) |
pdf |
| 12 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-2 | -5.913e+00 | 0.0555 | 79.0 | 7.00% | 73.8 | 5.11% | motif file (matrix) |
pdf |
| 13 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-2 | -5.815e+00 | 0.0565 | 76.0 | 6.74% | 70.6 | 4.89% | motif file (matrix) |
pdf |
| 14 | 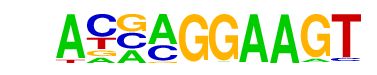 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-2 | -5.336e+00 | 0.0846 | 98.0 | 8.69% | 96.4 | 6.68% | motif file (matrix) |
pdf |
| 15 |  | HNF4a(NR/DR1)/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-2 | -4.824e+00 | 0.1318 | 56.0 | 4.96% | 51.6 | 3.58% | motif file (matrix) |
pdf |
| 16 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.730e+00 | 0.1357 | 110.0 | 9.75% | 112.3 | 7.78% | motif file (matrix) |
pdf |
| 17 |  | MafF(bZIP)/HepG2-MafF-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.707e+00 | 0.1357 | 35.0 | 3.10% | 29.1 | 2.01% | motif file (matrix) |
pdf |
| 18 |  | ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer | 1e-2 | -4.679e+00 | 0.1357 | 23.0 | 2.04% | 17.0 | 1.18% | motif file (matrix) |
pdf |



































