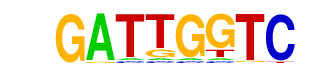
| p-value: | 1e-19 |
| log p-value: | -4.557e+01 |
| Information Content per bp: | 1.788 |
| Number of Target Sequences with motif | 22.0 |
| Percentage of Target Sequences with motif | 6.65% |
| Number of Background Sequences with motif | 1.3 |
| Percentage of Background Sequences with motif | 0.49% |
| Average Position of motif in Targets | 80.3 +/- 51.9bp |
| Average Position of motif in Background | 130.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | 1.8 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0502.1_NFYB/Jaspar
| Match Rank: | 1 |
| Score: | 0.77
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GACCAATC--
AAATGGACCAATCAG |
|


|
|
MA0060.2_NFYA/Jaspar
| Match Rank: | 2 |
| Score: | 0.76
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GACCAATC--------
TGGACCAATCAGCACTCT |
|


|
|
NFY(CCAAT)/Promoter/Homer
| Match Rank: | 3 |
| Score: | 0.72
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GACCAATC--
AGCCAATCGG |
|


|
|
POL004.1_CCAAT-box/Jaspar
| Match Rank: | 4 |
| Score: | 0.70
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GACCAATC-
ACTAGCCAATCA |
|


|
|
PB0144.1_Lef1_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.70
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GACCAATC-----
GAAGATCAATCACTTA |
|


|
|
PB0188.1_Tcf7l2_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.70
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GACCAATC-----
GAAGATCAATCACTAA |
|


|
|
MA0070.1_PBX1/Jaspar
| Match Rank: | 7 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GACCAATC---
CCATCAATCAAA |
|


|
|
MA0038.1_Gfi1/Jaspar
| Match Rank: | 8 |
| Score: | 0.66
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GACCAATC----
--CAAATCACTG |
|


|
|
MA0087.1_Sox5/Jaspar
| Match Rank: | 9 |
| Score: | 0.66
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GACCAATC
NAACAAT- |
|


|
|
HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer
| Match Rank: | 10 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GACCAATC-
DGATCRATAN |
|


|
|