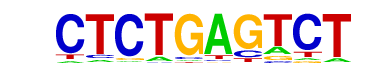
| p-value: | 1e-10 |
| log p-value: | -2.362e+01 |
| Information Content per bp: | 1.669 |
| Number of Target Sequences with motif | 22.0 |
| Percentage of Target Sequences with motif | 6.65% |
| Number of Background Sequences with motif | 3.9 |
| Percentage of Background Sequences with motif | 1.47% |
| Average Position of motif in Targets | 106.8 +/- 47.5bp |
| Average Position of motif in Background | 86.3 +/- 58.3bp |
| Strand Bias (log2 ratio + to - strand density) | -1.3 |
| Multiplicity (# of sites on avg that occur together) | 1.14 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0478.1_FOSL2/Jaspar
| Match Rank: | 1 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CTCTGAGTCT---
--NTGAGTCATCN |
|


|
|
MA0490.1_JUNB/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CTCTGAGTCT---
--ATGAGTCATCN |
|


|
|
MA0476.1_FOS/Jaspar
| Match Rank: | 3 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTCTGAGTCT--
-TGTGACTCATT |
|


|
|
MA0477.1_FOSL1/Jaspar
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CTCTGAGTCT--
-NATGAGTCACC |
|


|
|
MA0099.2_JUN::FOS/Jaspar
| Match Rank: | 5 |
| Score: | 0.60
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CTCTGAGTCT
---TGAGTCA |
|


|
|
AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CTCTGAGTCT-
-GATGAGTCAT |
|


|
|
BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer
| Match Rank: | 7 |
| Score: | 0.59
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTCTGAGTCT-
-DATGASTCAT |
|


|
|
Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer
| Match Rank: | 8 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTCTGAGTCT
TGCTGAGTCA |
|

|
|
Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTCTGAGTCT---
-DATGASTCATHN |
|


|
|
Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTCTGAGTCT---
-NATGACTCATNN |
|


|
|