| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Pax8(Paired/Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-8 | -1.871e+01 | 0.0000 | 12.0 | 3.63% | 0.8 | 0.29% | motif file (matrix) |
pdf |
| 2 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-5 | -1.213e+01 | 0.0007 | 156.0 | 47.13% | 94.7 | 35.46% | motif file (matrix) |
pdf |
| 3 |  | NFAT:AP1/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-4 | -1.149e+01 | 0.0007 | 12.0 | 3.63% | 2.9 | 1.10% | motif file (matrix) |
pdf |
| 4 |  | PAX5(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-4 | -1.149e+01 | 0.0007 | 12.0 | 3.63% | 2.0 | 0.75% | motif file (matrix) |
pdf |
| 5 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-4 | -1.137e+01 | 0.0007 | 97.0 | 29.31% | 52.5 | 19.67% | motif file (matrix) |
pdf |
| 6 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-4 | -9.868e+00 | 0.0021 | 11.0 | 3.32% | 2.7 | 1.00% | motif file (matrix) |
pdf |
| 7 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.333e+00 | 0.0085 | 10.0 | 3.02% | 2.5 | 0.93% | motif file (matrix) |
pdf |
| 8 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-3 | -8.139e+00 | 0.0085 | 7.0 | 2.11% | 1.3 | 0.48% | motif file (matrix) |
pdf |
| 9 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-3 | -8.139e+00 | 0.0085 | 7.0 | 2.11% | 1.1 | 0.40% | motif file (matrix) |
pdf |
| 10 |  | Srebp2(HLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.139e+00 | 0.0085 | 7.0 | 2.11% | 1.1 | 0.42% | motif file (matrix) |
pdf |
| 11 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-3 | -7.715e+00 | 0.0100 | 12.0 | 3.63% | 3.0 | 1.12% | motif file (matrix) |
pdf |
| 12 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-3 | -7.464e+00 | 0.0118 | 14.0 | 4.23% | 4.8 | 1.81% | motif file (matrix) |
pdf |
| 13 |  | Srebp1a(HLH)/HepG2-Srebp1a-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.891e+00 | 0.0192 | 9.0 | 2.72% | 2.4 | 0.91% | motif file (matrix) |
pdf |
| 14 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-2 | -6.730e+00 | 0.0210 | 21.0 | 6.34% | 8.9 | 3.33% | motif file (matrix) |
pdf |
| 15 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-2 | -6.551e+00 | 0.0234 | 66.0 | 19.94% | 37.8 | 14.14% | motif file (matrix) |
pdf |
| 16 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-2 | -6.455e+00 | 0.0242 | 42.0 | 12.69% | 21.6 | 8.10% | motif file (matrix) |
pdf |
| 17 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-2 | -6.368e+00 | 0.0248 | 6.0 | 1.81% | 1.1 | 0.41% | motif file (matrix) |
pdf |
| 18 |  | NFY(CCAAT)/Promoter/Homer | 1e-2 | -6.111e+00 | 0.0303 | 22.0 | 6.65% | 9.7 | 3.62% | motif file (matrix) |
pdf |
| 19 |  | STAT6/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-2 | -5.874e+00 | 0.0364 | 30.0 | 9.06% | 14.5 | 5.43% | motif file (matrix) |
pdf |
| 20 |  | Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer | 1e-2 | -5.455e+00 | 0.0526 | 94.0 | 28.40% | 59.1 | 22.12% | motif file (matrix) |
pdf |
| 21 | 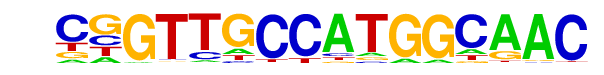 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-2 | -4.747e+00 | 0.1017 | 5.0 | 1.51% | 1.1 | 0.40% | motif file (matrix) |
pdf |
| 22 |  | bZIP:IRF/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-2 | -4.733e+00 | 0.1017 | 15.0 | 4.53% | 6.9 | 2.58% | motif file (matrix) |
pdf |
| 23 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-2 | -4.733e+00 | 0.1017 | 15.0 | 4.53% | 6.3 | 2.35% | motif file (matrix) |
pdf |













































