| p-value: | 1e-33 |
| log p-value: | -7.762e+01 |
| Information Content per bp: | 1.725 |
| Number of Target Sequences with motif | 33.0 |
| Percentage of Target Sequences with motif | 15.28% |
| Number of Background Sequences with motif | 2.0 |
| Percentage of Background Sequences with motif | 1.29% |
| Average Position of motif in Targets | 106.6 +/- 68.0bp |
| Average Position of motif in Background | 88.7 +/- 20.6bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.09 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
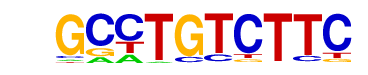
Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 1 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCCTGTCTTC
VBSYGTCTGG |
|

|
|
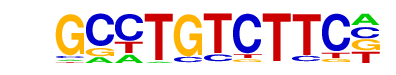
MA0480.1_Foxo1/Jaspar
| Match Rank: | 2 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCCTGTCTTC-
TCCTGTTTACA |
|

|
|
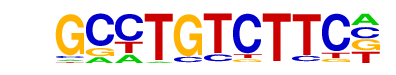
POL008.1_DCE_S_I/Jaspar
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | GCCTGTCTTC-
-----GCTTCC |
|

|
|
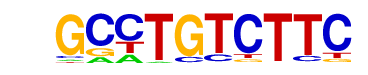
Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GCCTGTCTTC
--CTGTCTGG |
|

|
|
MA0157.1_FOXO3/Jaspar
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | GCCTGTCTTC-
---TGTTTACA |
|


|
|
Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GCCTGTCTTC
--CTGTTTAC |
|

|
|
Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer
| Match Rank: | 7 |
| Score: | 0.59
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GCCTGTCTTC
--TWGTCTGV |
|

|
|
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GCCTGTCTTC
-GCTGTG--- |
|

|
|
Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GCCTGTCTTC---
-SCTGTCARCACC |
|

|
|
Pax8(Paired/Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GCCTGTCTTC
GTCATGCHTGRCTGS |
|


|
|





















