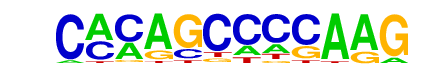
| p-value: | 1e-15 |
| log p-value: | -3.665e+01 |
| Information Content per bp: | 1.643 |
| Number of Target Sequences with motif | 20.0 |
| Percentage of Target Sequences with motif | 12.42% |
| Number of Background Sequences with motif | 2.7 |
| Percentage of Background Sequences with motif | 1.28% |
| Average Position of motif in Targets | 101.2 +/- 62.6bp |
| Average Position of motif in Background | 72.3 +/- 30.2bp |
| Strand Bias (log2 ratio + to - strand density) | 0.6 |
| Multiplicity (# of sites on avg that occur together) | 1.15 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0524.1_TFAP2C/Jaspar
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CACAGCCCCAAG---
CATGGCCCCAGGGCA |
|


|
|
MA0003.2_TFAP2A/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CACAGCCCCAAG---
CATTGCCTCAGGGCA |
|


|
|
AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 3 |
| Score: | 0.57
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CACAGCCCCAAG----
----GCCTCAGGGCAT |
|


|
|
AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer
| Match Rank: | 4 |
| Score: | 0.56
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | CACAGCCCCAAG----
----SCCTSAGGSCAW |
|


|
|
PB0088.1_Tcfap2e_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.55
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CACAGCCCCAAG----
-ATTGCCTGAGGCAAT |
|


|
|
MA0002.2_RUNX1/Jaspar
| Match Rank: | 6 |
| Score: | 0.54
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CACAGCCCCAAG-
--AAACCACAGAN |
|


|
|
PB0086.1_Tcfap2b_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.53
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CACAGCCCCAAG----
--TTGCCCTAGGGCAT |
|


|
|
MA0510.1_RFX5/Jaspar
| Match Rank: | 8 |
| Score: | 0.53
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CACAGCCCCAAG
CTCCCTGGCAACAGC |
|


|
|
ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 9 |
| Score: | 0.53
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CACAGCCCCAAG-
---ANCCGGAAGT |
|


|
|
PB0087.1_Tcfap2c_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CACAGCCCCAAG----
-ATTGCCTGAGGCGAA |
|


|
|