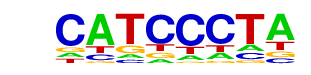
| p-value: | 1e-15 |
| log p-value: | -3.640e+01 |
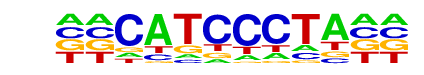
| Information Content per bp: | 1.474 |
| Number of Target Sequences with motif | 23.0 |
| Percentage of Target Sequences with motif | 14.29% |
| Number of Background Sequences with motif | 3.3 |
| Percentage of Background Sequences with motif | 1.53% |
| Average Position of motif in Targets | 97.6 +/- 45.0bp |
| Average Position of motif in Background | 131.2 +/- 27.1bp |
| Strand Bias (log2 ratio + to - strand density) | -1.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
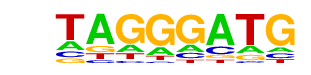
PB0098.1_Zfp410_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.69
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CATCCCTA----
NNNTCCATCCCATAANN |
|


|
|
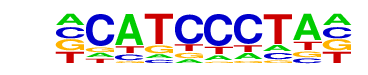
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer
| Match Rank: | 2 |
| Score: | 0.67
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CATCCCTA-
RCATTCCWGG |
|

|
|
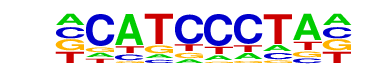
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 3 |
| Score: | 0.67
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CATCCCTA-
GCATTCCAGN |
|

|
|
MA0090.1_TEAD1/Jaspar
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CATCCCTA--
CACATTCCTCCG |
|

|
|
MA0136.1_ELF5/Jaspar
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CATCCCTA
TACTTCCTT- |
|


|
|
MA0056.1_MZF1_1-4/Jaspar
| Match Rank: | 6 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CATCCCTA
--TCCCCA |
|

|
|
POL002.1_INR/Jaspar
| Match Rank: | 7 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CATCCCTA
TCAGTCTT- |
|


|
|
PH0130.1_Otx2/Jaspar
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------CATCCCTA--
GANNATTAATCCCTNNN |
|


|
|
PH0129.1_Otx1/Jaspar
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------CATCCCTA--
NNNAATTAATCCCCNCN |
|


|
|
PH0137.1_Pitx1/Jaspar
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------CATCCCTA---
NTTGTTAATCCCTCTNN |
|


|
|