| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | STAT6(Stat)/CD4-Stat6-ChIP-Seq(GSE22104)/Homer | 1e-6 | -1.543e+01 | 0.0000 | 11.0 | 10.68% | 0.5 | 0.67% | motif file (matrix) |
pdf |
| 2 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-6 | -1.543e+01 | 0.0000 | 11.0 | 10.68% | 1.0 | 1.40% | motif file (matrix) |
pdf |
| 3 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-6 | -1.473e+01 | 0.0000 | 23.0 | 22.33% | 5.8 | 7.98% | motif file (matrix) |
pdf |
| 4 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-5 | -1.327e+01 | 0.0001 | 10.0 | 9.71% | 1.6 | 2.21% | motif file (matrix) |
pdf |
| 5 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-5 | -1.216e+01 | 0.0003 | 13.0 | 12.62% | 2.9 | 3.99% | motif file (matrix) |
pdf |
| 6 |  | STAT6/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-4 | -1.122e+01 | 0.0005 | 9.0 | 8.74% | 0.5 | 0.68% | motif file (matrix) |
pdf |
| 7 |  | NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-4 | -9.284e+00 | 0.0029 | 8.0 | 7.77% | 2.0 | 2.70% | motif file (matrix) |
pdf |
| 8 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-4 | -9.284e+00 | 0.0029 | 8.0 | 7.77% | 1.1 | 1.50% | motif file (matrix) |
pdf |
| 9 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-3 | -9.194e+00 | 0.0029 | 25.0 | 24.27% | 8.4 | 11.44% | motif file (matrix) |
pdf |
| 10 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-3 | -8.962e+00 | 0.0032 | 11.0 | 10.68% | 2.3 | 3.13% | motif file (matrix) |
pdf |
| 11 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.451e+00 | 0.0048 | 18.0 | 17.48% | 5.7 | 7.80% | motif file (matrix) |
pdf |
| 12 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-3 | -8.049e+00 | 0.0065 | 13.0 | 12.62% | 3.2 | 4.38% | motif file (matrix) |
pdf |
| 13 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-3 | -7.616e+00 | 0.0093 | 36.0 | 34.95% | 15.9 | 21.77% | motif file (matrix) |
pdf |
| 14 | 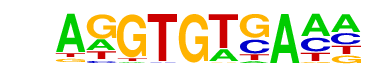 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-3 | -7.501e+00 | 0.0097 | 10.0 | 9.71% | 2.1 | 2.86% | motif file (matrix) |
pdf |
| 15 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.470e+00 | 0.0097 | 7.0 | 6.80% | 1.8 | 2.45% | motif file (matrix) |
pdf |
| 16 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.470e+00 | 0.0097 | 7.0 | 6.80% | 0.6 | 0.77% | motif file (matrix) |
pdf |
| 17 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-3 | -7.470e+00 | 0.0097 | 7.0 | 6.80% | 1.6 | 2.19% | motif file (matrix) |
pdf |
| 18 |  | n-Myc(HLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.470e+00 | 0.0097 | 7.0 | 6.80% | 0.4 | 0.60% | motif file (matrix) |
pdf |
| 19 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-3 | -7.470e+00 | 0.0097 | 7.0 | 6.80% | 0.6 | 0.87% | motif file (matrix) |
pdf |
| 20 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-3 | -7.276e+00 | 0.0097 | 19.0 | 18.45% | 7.0 | 9.52% | motif file (matrix) |
pdf |
| 21 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-2 | -6.362e+00 | 0.0202 | 16.0 | 15.53% | 5.8 | 7.91% | motif file (matrix) |
pdf |
| 22 |  | CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.140e+00 | 0.0241 | 9.0 | 8.74% | 2.3 | 3.16% | motif file (matrix) |
pdf |
| 23 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.793e+00 | 0.0300 | 6.0 | 5.83% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 24 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-2 | -5.793e+00 | 0.0300 | 6.0 | 5.83% | 1.6 | 2.12% | motif file (matrix) |
pdf |
| 25 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-2 | -5.793e+00 | 0.0300 | 6.0 | 5.83% | 1.6 | 2.12% | motif file (matrix) |
pdf |
| 26 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-2 | -5.505e+00 | 0.0385 | 19.0 | 18.45% | 7.4 | 10.09% | motif file (matrix) |
pdf |
| 27 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.887e+00 | 0.0663 | 8.0 | 7.77% | 2.2 | 2.96% | motif file (matrix) |
pdf |
| 28 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-2 | -4.887e+00 | 0.0663 | 8.0 | 7.77% | 2.7 | 3.74% | motif file (matrix) |
pdf |























































