
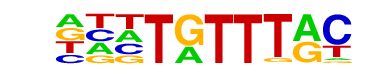
















| p-value: | 1e-1507 |
| log p-value: | -3.472e+03 |
| Information Content per bp: | 1.697 |
| Number of Target Sequences with motif | 10126.0 |
| Percentage of Target Sequences with motif | 23.51% |
| Number of Background Sequences with motif | 4157.0 |
| Percentage of Background Sequences with motif | 9.74% |
| Average Position of motif in Targets | 99.1 +/- 53.0bp |
| Average Position of motif in Background | 99.7 +/- 62.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.31 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.926 |  | 1e-1501 | -3457.738358 | 18.30% | 6.45% | motif file (matrix) |
| 2 | 0.984 |  | 1e-1406 | -3239.007584 | 24.37% | 10.69% | motif file (matrix) |
| 3 | 0.784 |  | 1e-823 | -1897.321707 | 22.68% | 12.04% | motif file (matrix) |
| 4 | 0.710 |  | 1e-780 | -1796.483697 | 18.89% | 9.44% | motif file (matrix) |
| 5 | 0.625 |  | 1e-485 | -1116.921685 | 18.89% | 11.13% | motif file (matrix) |
| 6 | 0.611 |  | 1e-482 | -1110.625892 | 14.07% | 7.45% | motif file (matrix) |
| 7 | 0.937 |  | 1e-397 | -914.671539 | 5.98% | 2.29% | motif file (matrix) |
| 8 | 0.763 |  | 1e-354 | -815.195008 | 15.12% | 9.07% | motif file (matrix) |
| 9 | 0.734 |  | 1e-343 | -791.911669 | 5.30% | 2.06% | motif file (matrix) |
| 10 | 0.685 |  | 1e-301 | -693.929327 | 4.76% | 1.87% | motif file (matrix) |
| 11 | 0.744 |  | 1e-266 | -614.626249 | 4.07% | 1.56% | motif file (matrix) |
| 12 | 0.629 |  | 1e-180 | -416.212641 | 0.80% | 0.10% | motif file (matrix) |
| 13 | 0.649 |  | 1e-178 | -411.252352 | 3.41% | 1.47% | motif file (matrix) |
| 14 | 0.761 |  | 1e-173 | -399.178658 | 5.97% | 3.29% | motif file (matrix) |
| 15 | 0.617 |  | 1e-130 | -299.640612 | 1.88% | 0.70% | motif file (matrix) |
| 16 | 0.620 |  | 1e-38 | -88.849156 | 0.14% | 0.02% | motif file (matrix) |