
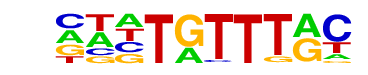













| p-value: | 1e-1465 |
| log p-value: | -3.374e+03 |
| Information Content per bp: | 1.611 |
| Number of Target Sequences with motif | 8127.0 |
| Percentage of Target Sequences with motif | 15.79% |
| Number of Background Sequences with motif | 2913.0 |
| Percentage of Background Sequences with motif | 5.71% |
| Average Position of motif in Targets | 99.7 +/- 51.7bp |
| Average Position of motif in Background | 100.1 +/- 59.9bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.19 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.932 |  | 1e-1429 | -3292.337555 | 19.36% | 8.06% | motif file (matrix) |
| 2 | 0.926 |  | 1e-1361 | -3133.838762 | 20.66% | 9.15% | motif file (matrix) |
| 3 | 0.922 |  | 1e-1066 | -2456.192676 | 15.81% | 6.82% | motif file (matrix) |
| 4 | 0.883 |  | 1e-1029 | -2370.463664 | 9.79% | 3.19% | motif file (matrix) |
| 5 | 0.848 |  | 1e-720 | -1659.992241 | 48.21% | 35.81% | motif file (matrix) |
| 6 | 0.738 |  | 1e-564 | -1298.861039 | 19.65% | 11.82% | motif file (matrix) |
| 7 | 0.600 |  | 1e-482 | -1111.417970 | 35.29% | 25.90% | motif file (matrix) |
| 8 | 0.724 |  | 1e-288 | -664.336326 | 6.85% | 3.53% | motif file (matrix) |
| 9 | 0.640 |  | 1e-230 | -529.645437 | 2.60% | 0.93% | motif file (matrix) |
| 10 | 0.632 |  | 1e-218 | -502.384523 | 6.45% | 3.58% | motif file (matrix) |
| 11 | 0.664 |  | 1e-157 | -362.458777 | 1.84% | 0.67% | motif file (matrix) |
| 12 | 0.728 |  | 1e-79 | -182.234466 | 2.42% | 1.35% | motif file (matrix) |
| 13 | 0.642 |  | 1e-70 | -163.421574 | 0.35% | 0.06% | motif file (matrix) |