















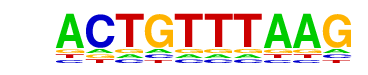


| p-value: | 1e-419 |
| log p-value: | -9.658e+02 |
| Information Content per bp: | 1.588 |
| Number of Target Sequences with motif | 3715.0 |
| Percentage of Target Sequences with motif | 26.76% |
| Number of Background Sequences with motif | 4596.5 |
| Percentage of Background Sequences with motif | 12.83% |
| Average Position of motif in Targets | 101.7 +/- 52.5bp |
| Average Position of motif in Background | 99.0 +/- 60.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.24 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.959 |  | 1e-380 | -876.973164 | 25.77% | 12.62% | motif file (matrix) |
| 2 | 0.972 |  | 1e-352 | -811.659210 | 18.42% | 7.82% | motif file (matrix) |
| 3 | 0.888 |  | 1e-279 | -643.967600 | 47.08% | 32.44% | motif file (matrix) |
| 4 | 0.893 |  | 1e-209 | -482.601229 | 20.81% | 11.60% | motif file (matrix) |
| 5 | 0.700 |  | 1e-148 | -342.597689 | 21.16% | 13.15% | motif file (matrix) |
| 6 | 0.767 |  | 1e-128 | -296.645044 | 34.27% | 25.06% | motif file (matrix) |
| 7 | 0.682 |  | 1e-127 | -294.555898 | 17.27% | 10.49% | motif file (matrix) |
| 8 | 0.674 |  | 1e-107 | -247.980488 | 12.88% | 7.48% | motif file (matrix) |
| 9 | 0.665 |  | 1e-87 | -200.573695 | 19.00% | 13.01% | motif file (matrix) |
| 10 | 0.726 |  | 1e-67 | -155.688280 | 2.28% | 0.72% | motif file (matrix) |
| 11 | 0.662 |  | 1e-67 | -155.392593 | 2.41% | 0.78% | motif file (matrix) |
| 12 | 0.717 |  | 1e-66 | -153.960961 | 4.14% | 1.84% | motif file (matrix) |
| 13 | 0.705 |  | 1e-57 | -132.372532 | 1.47% | 0.37% | motif file (matrix) |
| 14 | 0.711 |  | 1e-41 | -95.753808 | 1.69% | 0.60% | motif file (matrix) |
| 15 | 0.754 |  | 1e-30 | -69.246844 | 11.89% | 8.98% | motif file (matrix) |
| 16 | 0.609 |  | 1e-17 | -40.870969 | 1.82% | 1.00% | motif file (matrix) |
| 17 | 0.602 |  | 1e-11 | -26.592260 | 0.09% | 0.01% | motif file (matrix) |