













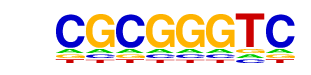




| p-value: | 1e-2361 |
| log p-value: | -5.437e+03 |
| Information Content per bp: | 1.850 |
| Number of Target Sequences with motif | 6975.0 |
| Percentage of Target Sequences with motif | 13.55% |
| Number of Background Sequences with motif | 800.2 |
| Percentage of Background Sequences with motif | 2.98% |
| Average Position of motif in Targets | 100.7 +/- 53.3bp |
| Average Position of motif in Background | 90.6 +/- 200.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.30 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.954 |  | 1e-1826 | -4206.755334 | 7.70% | 1.18% | motif file (matrix) |
| 2 | 0.740 |  | 1e-1531 | -3527.356392 | 2.35% | 0.05% | motif file (matrix) |
| 3 | 0.744 |  | 1e-1496 | -3446.543190 | 7.63% | 1.45% | motif file (matrix) |
| 4 | 0.936 |  | 1e-1327 | -3056.587856 | 6.08% | 1.02% | motif file (matrix) |
| 5 | 0.772 |  | 1e-1253 | -2885.201394 | 4.33% | 0.50% | motif file (matrix) |
| 6 | 0.697 |  | 1e-1215 | -2799.624326 | 27.55% | 14.75% | motif file (matrix) |
| 7 | 0.726 |  | 1e-945 | -2178.136987 | 11.83% | 4.61% | motif file (matrix) |
| 8 | 0.704 |  | 1e-944 | -2173.964450 | 1.15% | 0.01% | motif file (matrix) |
| 9 | 0.612 |  | 1e-848 | -1952.781272 | 1.06% | 0.01% | motif file (matrix) |
| 10 | 0.681 |  | 1e-750 | -1728.089350 | 0.88% | 0.01% | motif file (matrix) |
| 11 | 0.644 |  | 1e-737 | -1698.890036 | 9.18% | 3.53% | motif file (matrix) |
| 12 | 0.680 |  | 1e-704 | -1622.043225 | 1.20% | 0.04% | motif file (matrix) |
| 13 | 0.615 |  | 1e-430 | -991.812701 | 0.84% | 0.03% | motif file (matrix) |
| 14 | 0.801 |  | 1e-349 | -804.262877 | 1.58% | 0.26% | motif file (matrix) |
| 15 | 0.608 |  | 1e-327 | -753.298262 | 0.93% | 0.08% | motif file (matrix) |
| 16 | 0.710 |  | 1e-264 | -608.633770 | 1.04% | 0.14% | motif file (matrix) |
| 17 | 0.653 |  | 1e-164 | -378.937200 | 1.00% | 0.22% | motif file (matrix) |