
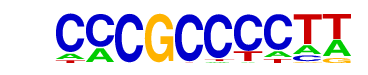














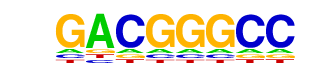


| p-value: | 1e-2662 |
| log p-value: | -6.131e+03 |
| Information Content per bp: | 1.774 |
| Number of Target Sequences with motif | 5486.0 |
| Percentage of Target Sequences with motif | 12.74% |
| Number of Background Sequences with motif | 432.3 |
| Percentage of Background Sequences with motif | 1.87% |
| Average Position of motif in Targets | 100.8 +/- 53.2bp |
| Average Position of motif in Background | 90.4 +/- 215.9bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.29 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.864 |  | 1e-2234 | -5144.079260 | 11.51% | 1.84% | motif file (matrix) |
| 2 | 0.816 |  | 1e-1551 | -3573.302456 | 13.38% | 3.61% | motif file (matrix) |
| 3 | 0.904 |  | 1e-1282 | -2952.741137 | 22.96% | 10.19% | motif file (matrix) |
| 4 | 0.757 |  | 1e-1115 | -2569.435359 | 1.44% | 0.01% | motif file (matrix) |
| 5 | 0.836 |  | 1e-1041 | -2397.278625 | 25.12% | 12.78% | motif file (matrix) |
| 6 | 0.787 |  | 1e-810 | -1867.060327 | 1.92% | 0.08% | motif file (matrix) |
| 7 | 0.759 |  | 1e-786 | -1809.981909 | 0.95% | 0.00% | motif file (matrix) |
| 8 | 0.643 |  | 1e-685 | -1579.567885 | 1.40% | 0.04% | motif file (matrix) |
| 9 | 0.681 |  | 1e-475 | -1095.403713 | 0.96% | 0.03% | motif file (matrix) |
| 10 | 0.622 |  | 1e-392 | -904.238089 | 0.91% | 0.04% | motif file (matrix) |
| 11 | 0.773 |  | 1e-386 | -889.677362 | 1.86% | 0.27% | motif file (matrix) |
| 12 | 0.654 |  | 1e-381 | -878.022594 | 0.62% | 0.01% | motif file (matrix) |
| 13 | 0.685 |  | 1e-355 | -818.954029 | 2.39% | 0.49% | motif file (matrix) |
| 14 | 0.638 |  | 1e-344 | -793.292036 | 0.49% | 0.00% | motif file (matrix) |
| 15 | 0.786 |  | 1e-310 | -715.610814 | 1.70% | 0.28% | motif file (matrix) |
| 16 | 0.727 |  | 1e-277 | -638.031196 | 0.65% | 0.03% | motif file (matrix) |
| 17 | 0.674 |  | 1e-115 | -266.664321 | 0.45% | 0.05% | motif file (matrix) |