| p-value: | 1e-788 |
| log p-value: | -1.815e+03 |
| Information Content per bp: | 1.794 |
| Number of Target Sequences with motif | 470.0 |
| Percentage of Target Sequences with motif | 1.09% |
| Number of Background Sequences with motif | 2.1 |
| Percentage of Background Sequences with motif | 0.01% |
| Average Position of motif in Targets | 93.9 +/- 53.5bp |
| Average Position of motif in Background | 164.3 +/- 4.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0506.1_NRF1/Jaspar
| Match Rank: | 1 |
| Score: | 0.79
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TGCGGCCGCGCG
-GCGCCTGCGCA |
|


|
|
NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 2 |
| Score: | 0.73
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGCGGCCGCGCG
CTGCGCATGCGC- |
|


|
|
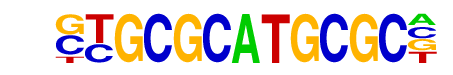
NRF1/Promoter/Homer
| Match Rank: | 3 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGCGGCCGCGCG
GTGCGCATGCGC- |
|

|
|
MA0048.1_NHLH1/Jaspar
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGCGGCCGCGCG
NCGCAGCTGCGN- |
|


|
|
PB0095.1_Zfp161_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TGCGGCCGCGCG-----
-NCANGCGCGCGCGCCA |
|


|
|
POL011.1_XCPE1/Jaspar
| Match Rank: | 6 |
| Score: | 0.56
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TGCGGCCGCGCG
-GGTCCCGCCC- |
|


|
|
PB0010.1_Egr1_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TGCGGCCGCGCG--
TCCGCCCCCGCATT |
|


|
|
Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer
| Match Rank: | 8 |
| Score: | 0.54
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCGGCCGCGCG
YCCGCCCACGCN |
|


|
|
MA0469.1_E2F3/Jaspar
| Match Rank: | 9 |
| Score: | 0.53
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGCGGCCGCGCG--
NNGTGNGGGCGGGAG |
|


|
|
MA0162.2_EGR1/Jaspar
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCGGCCGCGCG--
GGCGGGGGCGGGGG |
|


|
|





















