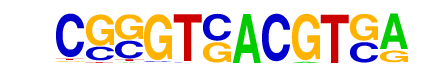








| p-value: | 1e-577 |
| log p-value: | -1.329e+03 |
| Information Content per bp: | 1.432 |
| Number of Target Sequences with motif | 604.0 |
| Percentage of Target Sequences with motif | 1.40% |
| Number of Background Sequences with motif | 14.1 |
| Percentage of Background Sequences with motif | 0.06% |
| Average Position of motif in Targets | 101.0 +/- 54.3bp |
| Average Position of motif in Background | 105.4 +/- 45.5bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.636 |  | 1e-568 | -1308.424163 | 0.73% | 0.01% | motif file (matrix) |
| 2 | 0.606 |  | 1e-520 | -1199.569910 | 0.79% | 0.01% | motif file (matrix) |
| 3 | 0.632 |  | 1e-500 | -1151.832801 | 1.27% | 0.06% | motif file (matrix) |
| 4 | 0.638 |  | 1e-475 | -1095.740798 | 5.77% | 1.94% | motif file (matrix) |
| 5 | 0.649 |  | 1e-254 | -586.520002 | 0.58% | 0.03% | motif file (matrix) |
| 6 | 0.627 |  | 1e-207 | -477.190957 | 1.12% | 0.18% | motif file (matrix) |
| 7 | 0.618 |  | 1e-143 | -330.668179 | 0.46% | 0.04% | motif file (matrix) |