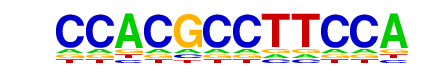
| p-value: | 1e-44 |
| log p-value: | -1.025e+02 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 40.0 |
| Percentage of Target Sequences with motif | 0.22% |
| Number of Background Sequences with motif | 1.9 |
| Percentage of Background Sequences with motif | 0.01% |
| Average Position of motif in Targets | 95.4 +/- 49.8bp |
| Average Position of motif in Background | 124.8 +/- 27.4bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0180.1_Sp4_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.73
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TGGAAGGCGTGG----
-CAAAGGCGTGGCCAG |
|


|
|
MA0006.1_Arnt::Ahr/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | TGGAAGGCGTGG
-----TGCGTG- |
|


|
|
KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer
| Match Rank: | 3 |
| Score: | 0.60
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | TGGAAGGCGTGG-
---DGGGYGKGGC |
|


|
|
PB0039.1_Klf7_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGGAAGGCGTGG----
NNAGGGGCGGGGTNNA |
|


|
|
ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | TGGAAGGCGTGG
----AGGCCTNG |
|


|
|
POL006.1_BREu/Jaspar
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | TGGAAGGCGTGG-
-----GGCGCGCT |
|


|
|
MA0039.2_Klf4/Jaspar
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | TGGAAGGCGTGG-
---TGGGTGGGGC |
|


|
|
MA0493.1_Klf1/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TGGAAGGCGTGG--
---TGGGTGTGGCN |
|


|
|
MA0598.1_EHF/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGGAAGGCGTGG
CAGGAAGG----- |
|


|
|
PB0110.1_Bcl6b_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGGAAGGCGTGG----
NNTNAGGGGCGGNNNN |
|


|
|