

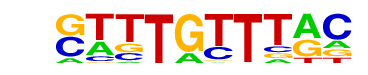









| p-value: | 1e-396 |
| log p-value: | -9.124e+02 |
| Information Content per bp: | 1.654 |
| Number of Target Sequences with motif | 2958.0 |
| Percentage of Target Sequences with motif | 21.31% |
| Number of Background Sequences with motif | 995.3 |
| Percentage of Background Sequences with motif | 9.27% |
| Average Position of motif in Targets | 100.8 +/- 53.2bp |
| Average Position of motif in Background | 102.0 +/- 62.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.18 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.899 |  | 1e-377 | -868.452622 | 19.03% | 7.95% | motif file (matrix) |
| 2 | 0.904 |  | 1e-348 | -802.255420 | 16.04% | 6.32% | motif file (matrix) |
| 3 | 0.892 |  | 1e-257 | -593.300989 | 30.64% | 18.54% | motif file (matrix) |
| 4 | 0.628 |  | 1e-155 | -358.622497 | 28.34% | 19.00% | motif file (matrix) |
| 5 | 0.685 |  | 1e-107 | -246.713809 | 11.10% | 6.14% | motif file (matrix) |
| 6 | 0.629 |  | 1e-70 | -163.361097 | 0.50% | 0.02% | motif file (matrix) |
| 7 | 0.680 |  | 1e-67 | -155.584811 | 0.40% | 0.01% | motif file (matrix) |
| 8 | 0.725 |  | 1e-50 | -116.475302 | 5.54% | 3.09% | motif file (matrix) |
| 9 | 0.641 |  | 1e-36 | -84.450360 | 0.25% | 0.01% | motif file (matrix) |
| 10 | 0.773 |  | 1e-29 | -67.336490 | 0.41% | 0.06% | motif file (matrix) |