














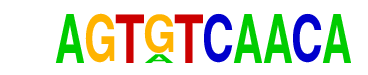


| p-value: | 1e-743 |
| log p-value: | -1.712e+03 |
| Information Content per bp: | 1.594 |
| Number of Target Sequences with motif | 4319.0 |
| Percentage of Target Sequences with motif | 18.78% |
| Number of Background Sequences with motif | 1886.8 |
| Percentage of Background Sequences with motif | 7.06% |
| Average Position of motif in Targets | 99.2 +/- 53.5bp |
| Average Position of motif in Background | 100.9 +/- 60.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.28 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.932 |  | 1e-672 | -1548.974917 | 19.26% | 7.79% | motif file (matrix) |
| 2 | 0.945 |  | 1e-429 | -990.074177 | 13.43% | 5.58% | motif file (matrix) |
| 3 | 0.678 |  | 1e-321 | -740.137456 | 31.78% | 20.94% | motif file (matrix) |
| 4 | 0.883 |  | 1e-245 | -565.286942 | 32.41% | 22.75% | motif file (matrix) |
| 5 | 0.766 |  | 1e-192 | -442.525299 | 11.20% | 6.03% | motif file (matrix) |
| 6 | 0.730 |  | 1e-148 | -342.948618 | 7.59% | 3.86% | motif file (matrix) |
| 7 | 0.663 |  | 1e-119 | -275.116222 | 5.68% | 2.80% | motif file (matrix) |
| 8 | 0.652 |  | 1e-118 | -272.397831 | 10.01% | 6.05% | motif file (matrix) |
| 9 | 0.679 |  | 1e-112 | -258.486660 | 2.21% | 0.67% | motif file (matrix) |
| 10 | 0.624 |  | 1e-105 | -243.632323 | 0.36% | 0.01% | motif file (matrix) |
| 11 | 0.747 |  | 1e-103 | -238.242415 | 4.94% | 2.44% | motif file (matrix) |
| 12 | 0.650 |  | 1e-99 | -228.986891 | 6.71% | 3.77% | motif file (matrix) |
| 13 | 0.692 |  | 1e-83 | -192.149282 | 4.70% | 2.47% | motif file (matrix) |
| 14 | 0.653 |  | 1e-55 | -128.024599 | 0.25% | 0.01% | motif file (matrix) |
| 15 | 0.650 |  | 1e-30 | -71.099326 | 0.24% | 0.03% | motif file (matrix) |
| 16 | 0.648 |  | 1e-28 | -66.751054 | 0.22% | 0.03% | motif file (matrix) |