



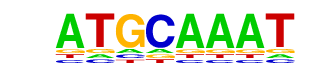




| p-value: | 1e-452 |
| log p-value: | -1.041e+03 |
| Information Content per bp: | 1.666 |
| Number of Target Sequences with motif | 3305.0 |
| Percentage of Target Sequences with motif | 14.64% |
| Number of Background Sequences with motif | 1673.3 |
| Percentage of Background Sequences with motif | 6.16% |
| Average Position of motif in Targets | 99.8 +/- 53.8bp |
| Average Position of motif in Background | 95.7 +/- 57.4bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.14 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.888 |  | 1e-265 | -610.919280 | 18.41% | 10.62% | motif file (matrix) |
| 2 | 0.820 |  | 1e-226 | -521.636464 | 10.08% | 4.85% | motif file (matrix) |
| 3 | 0.600 |  | 1e-170 | -392.698566 | 6.23% | 2.72% | motif file (matrix) |
| 4 | 0.788 |  | 1e-82 | -190.843841 | 2.66% | 1.08% | motif file (matrix) |
| 5 | 0.671 |  | 1e-66 | -152.101768 | 1.31% | 0.40% | motif file (matrix) |
| 6 | 0.662 |  | 1e-46 | -107.939058 | 0.20% | 0.01% | motif file (matrix) |
| 7 | 0.656 |  | 1e-35 | -81.388746 | 0.22% | 0.02% | motif file (matrix) |