| p-value: | 1e-54 |
| log p-value: | -1.245e+02 |
| Information Content per bp: | 1.683 |
| Number of Target Sequences with motif | 57.0 |
| Percentage of Target Sequences with motif | 0.59% |
| Number of Background Sequences with motif | 2.3 |
| Percentage of Background Sequences with motif | 0.03% |
| Average Position of motif in Targets | 95.7 +/- 50.5bp |
| Average Position of motif in Background | 125.4 +/- 50.7bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
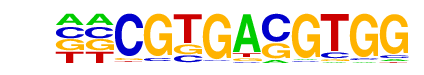
MF0002.1_bZIP_CREB/G-box-like_subclass/Jaspar
| Match Rank: | 1 |
| Score: | 0.70
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CGTGACGTGG
--TGACGT-- |
|


|
|
USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGTGACGTGG
GGTCACGTGA |
|


|
|
ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGTGACGTGG-
ACAGGATGTGGT |
|


|
|
MA0067.1_Pax2/Jaspar
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CGTGACGTGG
NCGTGACN--- |
|


|
|
NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 5 |
| Score: | 0.62
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGTGACGTGG-
-GTCACGTGGM |
|


|
|
MA0130.1_ZNF354C/Jaspar
| Match Rank: | 6 |
| Score: | 0.62
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | CGTGACGTGG--
------GTGGAT |
|


|
|
MA0104.3_Mycn/Jaspar
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CGTGACGTGG-
---CACGTGGC |
|


|
|
MA0259.1_HIF1A::ARNT/Jaspar
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CGTGACGTGG
--GGACGTGC |
|


|
|
CRE(bZIP)/Promoter/Homer
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGTGACGTGG-
CGGTGACGTCAC |
|


|
|
E-box(HLH)/Promoter/Homer
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CGTGACGTGG
CCGGTCACGTGA |
|

|
|





















