





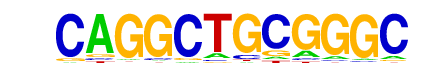




| p-value: | 1e-163 |
| log p-value: | -3.773e+02 |
| Information Content per bp: | 1.682 |
| Number of Target Sequences with motif | 128.0 |
| Percentage of Target Sequences with motif | 1.34% |
| Number of Background Sequences with motif | 2.7 |
| Percentage of Background Sequences with motif | 0.04% |
| Average Position of motif in Targets | 113.7 +/- 58.6bp |
| Average Position of motif in Background | 80.2 +/- 11.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.6 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.684 |  | 1e-150 | -346.250671 | 1.25% | 0.03% | motif file (matrix) |
| 2 | 0.632 |  | 1e-120 | -278.373610 | 1.62% | 0.12% | motif file (matrix) |
| 3 | 0.729 |  | 1e-110 | -254.157172 | 0.84% | 0.02% | motif file (matrix) |
| 4 | 0.637 |  | 1e-99 | -229.592518 | 0.77% | 0.02% | motif file (matrix) |
| 5 | 0.636 |  | 1e-92 | -213.484331 | 0.73% | 0.03% | motif file (matrix) |
| 6 | 0.668 |  | 1e-82 | -190.355880 | 0.91% | 0.05% | motif file (matrix) |
| 7 | 0.604 |  | 1e-70 | -162.785266 | 0.59% | 0.01% | motif file (matrix) |
| 8 | 0.708 |  | 1e-69 | -159.000111 | 0.58% | 0.01% | motif file (matrix) |
| 9 | 0.836 |  | 1e-43 | -100.407803 | 1.40% | 0.31% | motif file (matrix) |