
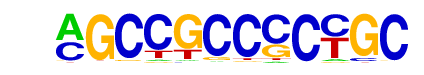


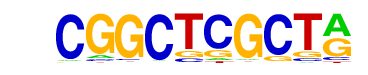

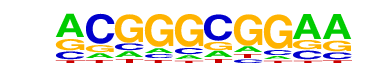







| p-value: | 1e-156 |
| log p-value: | -3.594e+02 |
| Information Content per bp: | 1.741 |
| Number of Target Sequences with motif | 211.0 |
| Percentage of Target Sequences with motif | 2.20% |
| Number of Background Sequences with motif | 12.6 |
| Percentage of Background Sequences with motif | 0.17% |
| Average Position of motif in Targets | 95.7 +/- 57.8bp |
| Average Position of motif in Background | 117.9 +/- 65.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.670 |  | 1e-137 | -316.180995 | 1.30% | 0.05% | motif file (matrix) |
| 2 | 0.705 |  | 1e-117 | -270.786296 | 0.88% | 0.03% | motif file (matrix) |
| 3 | 0.644 |  | 1e-103 | -237.728489 | 0.79% | 0.00% | motif file (matrix) |
| 4 | 0.652 |  | 1e-101 | -233.653831 | 0.78% | 0.02% | motif file (matrix) |
| 5 | 0.602 |  | 1e-96 | -221.510638 | 0.75% | 0.02% | motif file (matrix) |
| 6 | 0.710 |  | 1e-92 | -212.458372 | 0.98% | 0.05% | motif file (matrix) |
| 7 | 0.715 |  | 1e-78 | -180.897189 | 0.96% | 0.07% | motif file (matrix) |
| 8 | 0.651 |  | 1e-66 | -152.195053 | 1.04% | 0.11% | motif file (matrix) |
| 9 | 0.615 |  | 1e-64 | -147.752669 | 0.55% | 0.02% | motif file (matrix) |
| 10 | 0.607 |  | 1e-64 | -147.615184 | 5.65% | 2.51% | motif file (matrix) |
| 11 | 0.610 |  | 1e-55 | -127.643318 | 0.69% | 0.04% | motif file (matrix) |
| 12 | 0.661 |  | 1e-33 | -76.675555 | 0.49% | 0.05% | motif file (matrix) |