











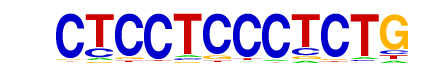


| p-value: | 1e-509 |
| log p-value: | -1.174e+03 |
| Information Content per bp: | 1.822 |
| Number of Target Sequences with motif | 377.0 |
| Percentage of Target Sequences with motif | 9.90% |
| Number of Background Sequences with motif | 4.7 |
| Percentage of Background Sequences with motif | 0.20% |
| Average Position of motif in Targets | 101.2 +/- 51.7bp |
| Average Position of motif in Background | 61.2 +/- 30.2bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.25 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.823 |  | 1e-397 | -915.982681 | 6.04% | 0.00% | motif file (matrix) |
| 2 | 0.795 |  | 1e-359 | -826.872529 | 5.57% | 0.02% | motif file (matrix) |
| 3 | 0.748 |  | 1e-310 | -715.380321 | 4.96% | 0.07% | motif file (matrix) |
| 4 | 0.614 |  | 1e-164 | -378.783491 | 3.02% | 0.04% | motif file (matrix) |
| 5 | 0.636 |  | 1e-133 | -307.538217 | 2.57% | 0.08% | motif file (matrix) |
| 6 | 0.734 |  | 1e-128 | -295.264832 | 2.49% | 0.08% | motif file (matrix) |
| 7 | 0.682 |  | 1e-126 | -292.343417 | 5.12% | 0.51% | motif file (matrix) |
| 8 | 0.702 |  | 1e-121 | -279.051174 | 2.39% | 0.09% | motif file (matrix) |
| 9 | 0.788 |  | 1e-118 | -272.815130 | 27.41% | 13.20% | motif file (matrix) |
| 10 | 0.639 |  | 1e-114 | -263.016667 | 2.28% | 0.08% | motif file (matrix) |
| 11 | 0.696 |  | 1e-82 | -188.984230 | 6.80% | 1.61% | motif file (matrix) |
| 12 | 0.601 |  | 1e-80 | -184.702930 | 2.65% | 0.22% | motif file (matrix) |
| 13 | 0.640 |  | 1e-63 | -146.333333 | 2.47% | 0.24% | motif file (matrix) |