| p-value: | 1e-49 |
| log p-value: | -1.128e+02 |
| Information Content per bp: | 1.509 |
| Number of Target Sequences with motif | 66.0 |
| Percentage of Target Sequences with motif | 1.73% |
| Number of Background Sequences with motif | 3.7 |
| Percentage of Background Sequences with motif | 0.16% |
| Average Position of motif in Targets | 99.9 +/- 53.7bp |
| Average Position of motif in Background | 124.2 +/- 32.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
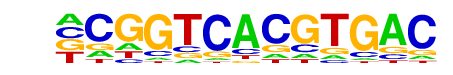
E-box(HLH)/Promoter/Homer
| Match Rank: | 1 |
| Score: | 0.90
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGGTCACGTGAC
CCGGTCACGTGA- |
|

|
|
MA0093.2_USF1/Jaspar
| Match Rank: | 2 |
| Score: | 0.87
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGGTCACGTGAC
-GGTCACGTGGC |
|


|
|
MA0526.1_USF2/Jaspar
| Match Rank: | 3 |
| Score: | 0.86
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGGTCACGTGAC
-GGTCACATGAC |
|


|
|
USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 4 |
| Score: | 0.84
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CGGTCACGTGAC
-GGTCACGTGA- |
|


|
|
PB0007.1_Bhlhb2_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.83
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CGGTCACGTGAC------
GGAAGAGTCACGTGACCAATAC |
|


|
|
Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer
| Match Rank: | 6 |
| Score: | 0.78
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CGGTCACGTGAC
--GTCACGTGGT |
|


|
|
CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 7 |
| Score: | 0.77
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CGGTCACGTGAC
--GHCACGTG-- |
|


|
|
bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 8 |
| Score: | 0.76
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGGTCACGTGAC
-NGKCACGTGM- |
|


|
|
BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 9 |
| Score: | 0.76
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CGGTCACGTGAC
--GNCACGTG-- |
|


|
|
NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 10 |
| Score: | 0.76
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CGGTCACGTGAC
--KCCACGTGAC |
|


|
|





















