


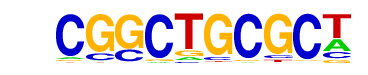



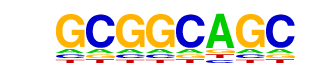

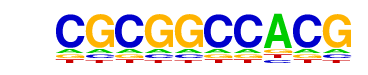


| p-value: | 1e-208 |
| log p-value: | -4.792e+02 |
| Information Content per bp: | 1.596 |
| Number of Target Sequences with motif | 138.0 |
| Percentage of Target Sequences with motif | 3.62% |
| Number of Background Sequences with motif | 1.9 |
| Percentage of Background Sequences with motif | 0.08% |
| Average Position of motif in Targets | 103.8 +/- 53.9bp |
| Average Position of motif in Background | 111.3 +/- 3.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.08 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.658 |  | 1e-206 | -474.776976 | 3.60% | 0.04% | motif file (matrix) |
| 2 | 0.637 |  | 1e-155 | -359.001443 | 3.44% | 0.13% | motif file (matrix) |
| 3 | 0.755 |  | 1e-146 | -336.877904 | 3.28% | 0.11% | motif file (matrix) |
| 4 | 0.660 |  | 1e-135 | -311.849391 | 5.33% | 0.52% | motif file (matrix) |
| 5 | 0.630 |  | 1e-135 | -311.650321 | 2.60% | 0.08% | motif file (matrix) |
| 6 | 0.693 |  | 1e-129 | -297.476893 | 4.17% | 0.29% | motif file (matrix) |
| 7 | 0.655 |  | 1e-122 | -282.898583 | 2.89% | 0.13% | motif file (matrix) |
| 8 | 0.712 |  | 1e-85 | -197.759344 | 2.23% | 0.12% | motif file (matrix) |
| 9 | 0.726 |  | 1e-72 | -167.429095 | 1.63% | 0.08% | motif file (matrix) |
| 10 | 0.794 |  | 1e-64 | -147.945449 | 2.07% | 0.15% | motif file (matrix) |